8IYM
 
 | | Crystal structure of a protein acetyltransferase, HP0935 | | Descriptor: | 1,2-ETHANEDIOL, N-acetyltransferase domain-containing protein, POTASSIUM ION, ... | | Authors: | Dadireddy, V, Mahanta, P, Kumar, A, Desirazu, R.N, Ramakumar, S. | | Deposit date: | 2023-04-05 | | Release date: | 2024-04-10 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structure of a protein acetyltransferase, HP0935
To be published
|
|
8IYO
 
 | | Crystal structure of a protein acetyltransferase, HP0935, acetyl-CoA bound form | | Descriptor: | ACETYL COENZYME *A, N-acetyltransferase domain-containing protein | | Authors: | Dadireddy, V, Mahanta, P, Kumar, A, Desirazu, R.N, Ramakumar, S. | | Deposit date: | 2023-04-05 | | Release date: | 2024-04-10 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Crystal structure of a protein acetyltransferase, HP0935, acetyl-CoA bound form
To be published
|
|
8HKR
 
 | | Crystal Structure of Histone H3 Lysine 79 (H3K79) Methyltransferase Rv2067c from Mycobacterium tuberculosis | | Descriptor: | PHOSPHATE ION, Protein lysine methyltransferase, S-ADENOSYL-L-HOMOCYSTEINE | | Authors: | Dadireddy, V, Singh, P.R, Kalladi, S.M, Valakunja, N, Ramakumar, S. | | Deposit date: | 2022-11-28 | | Release date: | 2023-10-18 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | The Mycobacterium tuberculosis methyltransferase Rv2067c manipulates host epigenetic programming to promote its own survival.
Nat Commun, 14, 2023
|
|
5H3L
 
 | | Structure of methylglyoxal synthase crystallised as a contaminant | | Descriptor: | FORMIC ACID, Methylglyoxal synthase | | Authors: | Hatti, K, Dadireddy, V, Srinivasan, N, Ramakumar, S, Murthy, M.R.N. | | Deposit date: | 2016-10-25 | | Release date: | 2016-11-09 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structure determination of contaminant proteins using the MarathonMR procedure.
J. Struct. Biol., 197, 2017
|
|
5GO6
 
 | | Structure of sortase E T196V mutant from Streptomyces avermitilis | | Descriptor: | Putative secreted protein, SULFATE ION | | Authors: | Das, S, Pawale, V.S, Dadireddy, V, Roy, R.P, Ramakumar, S. | | Deposit date: | 2016-07-26 | | Release date: | 2017-07-26 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structure of sortase E T196V mutant from Streptomyces avermitilis
To Be Published
|
|
5GO5
 
 | | Structure of sortase E from Streptomyces avermitilis | | Descriptor: | GLYCINE, SULFATE ION, sortase | | Authors: | Das, S, Pawale, V.S, Dadireddy, V, Roy, R.P, Ramakumar, S. | | Deposit date: | 2016-07-26 | | Release date: | 2017-03-15 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Structure and specificity of a new class of Ca2+-independent housekeeping sortase from Streptomyces avermitilis provide insights into its non-canonical substrate preference.
J. Biol. Chem., 292, 2017
|
|
5TX1
 
 | | Cryo-Electron microscopy structure of species-D human adenovirus 26 | | Descriptor: | Fiber, Hexon protein, PIIIa, ... | | Authors: | Reddy, V, Yu, X, Veesler, D. | | Deposit date: | 2016-11-15 | | Release date: | 2017-05-31 | | Last modified: | 2024-03-13 | | Method: | ELECTRON MICROSCOPY (3.7 Å) | | Cite: | Cryo-EM structure of human adenovirus D26 reveals the conservation of structural organization among human adenoviruses.
Sci Adv, 3, 2017
|
|
3COB
 
 | | Structural Dynamics of the Microtubule binding and regulatory elements in the Kinesin-like Calmodulin binding protein | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Kinesin heavy chain-like protein, MAGNESIUM ION | | Authors: | Vinogradova, M.V, Malanina, G.G, Reddy, V, Reddy, A.S.N, Fletterick, R.J. | | Deposit date: | 2008-03-27 | | Release date: | 2008-06-24 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structural dynamics of the microtubule binding and regulatory elements in the kinesin-like calmodulin binding protein.
J.Struct.Biol., 163, 2008
|
|
3CNZ
 
 | | Structural dynamics of the microtubule binding and regulatory elements in the kinesin-like calmodulin binding protein | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Kinesin heavy chain-like protein, MAGNESIUM ION | | Authors: | Vinogradova, M.V, Malanina, G.G, Reddy, V, Reddy, A.S.N, Fletterick, R.J. | | Deposit date: | 2008-03-26 | | Release date: | 2008-06-17 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Structural dynamics of the microtubule binding and regulatory elements in the kinesin-like calmodulin binding protein.
J.Struct.Biol., 163, 2008
|
|
1IH5
 
 | | CRYSTAL STRUCTURE OF AQUAPORIN-1 | | Descriptor: | AQUAPORIN-1 | | Authors: | Ren, G, Reddy, V.S, Cheng, A, Melnyk, P, Mitra, A.K. | | Deposit date: | 2001-04-18 | | Release date: | 2001-04-25 | | Last modified: | 2024-02-07 | | Method: | ELECTRON CRYSTALLOGRAPHY (3.7 Å) | | Cite: | Visualization of a water-selective pore by electron crystallography in vitreous ice.
Proc.Natl.Acad.Sci.USA, 98, 2001
|
|
1OHF
 
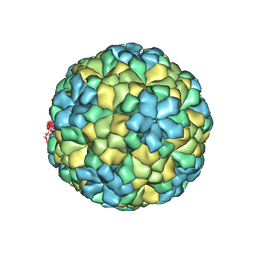 | | The refined structure of Nudaurelia capensis omega virus | | Descriptor: | MAGNESIUM ION, p70 | | Authors: | Helgstrand, C, Munshi, S, Johnson, J.E, Liljas, L. | | Deposit date: | 2003-05-26 | | Release date: | 2004-02-26 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | The Refined Structure of Nudaurelia Capensis Omega Virus Reveals Control Elements for a T = 4 Capsid Maturation
Virology, 318, 2004
|
|
8DEA
 
 | |
8DG6
 
 | |
8D95
 
 | |
7N7X
 
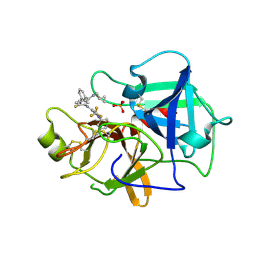 | | Crystal structure of BCX7353(ORLADEYO) in complex with human plasma kallikrein serine protease domain at 2.1 angstrom resolution | | Descriptor: | Orladeyo, PHOSPHATE ION, Plasma kallikrein light chain | | Authors: | Krishnan, R, Yarlagadda, B.S, Kotian, P, Polach, K.J, Zhang, W. | | Deposit date: | 2021-06-11 | | Release date: | 2021-09-15 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.097 Å) | | Cite: | Berotralstat (BCX7353): Structure-Guided Design of a Potent, Selective, and Oral Plasma Kallikrein Inhibitor to Prevent Attacks of Hereditary Angioedema (HAE).
J.Med.Chem., 64, 2021
|
|
5H4G
 
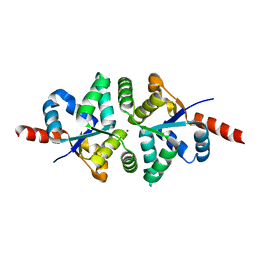 | | Structure of PIN-domain protein (VapC4 toxin) from Pyrococcus horikoshii determined at 1.77 A resolution | | Descriptor: | Ribonuclease VapC4, ZINC ION | | Authors: | Biswas, A, Hatti, K, Srinivasan, N, Murthy, M.R.N, Sekar, K. | | Deposit date: | 2016-10-31 | | Release date: | 2016-11-23 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.77 Å) | | Cite: | Structure determination of contaminant proteins using the MarathonMR procedure
J. Struct. Biol., 197, 2017
|
|
5H4H
 
 | | Structure of PIN-domain protein (VapC4 toxin) from Pyrococcus horikoshii determined at 2.2 A resolution | | Descriptor: | CADMIUM ION, Ribonuclease VapC4 | | Authors: | Biswas, A, Hatti, K, Srinivasan, N, Murthy, M.R.N, Sekar, K. | | Deposit date: | 2016-10-31 | | Release date: | 2016-11-23 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.23 Å) | | Cite: | Structure determination of contaminant proteins using the MarathonMR procedure
J. Struct. Biol., 197, 2017
|
|
5H4F
 
 | | Structure of inorganic pyrophosphatase crystallised as a contaminant | | Descriptor: | ZINC ION, inorganic pyrophosphatase | | Authors: | Chaudhary, S, Hatti, K, Srinivasan, N, Murthy, M.R.N, Sekar, K. | | Deposit date: | 2016-10-31 | | Release date: | 2016-11-16 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Structure determination of contaminant proteins using the MarathonMR procedure.
J. Struct. Biol., 197, 2017
|
|
