2KVJ
 
 | | NMR and MD solution structure of a Gamma-Methylated PNA duplex | | Descriptor: | Gamma-Modified Peptide Nucleic Acid | | Authors: | He, W, Crawford, M.J, Rapireddy, S, Madrid, M, Gil, R.R, Ly, D.H, Achim, C. | | Deposit date: | 2010-03-15 | | Release date: | 2010-05-12 | | Last modified: | 2023-11-15 | | Method: | SOLUTION NMR | | Cite: | The structure of a gamma-modified peptide nucleic acid duplex.
Mol Biosyst, 6, 2010
|
|
2DQ5
 
 | | solution structure of the Mid1 B Box2 Chc(D/C)C2H2 Zinc-Binding Domain: insights into an evolutionary conserved ring fold | | Descriptor: | Midline-1, ZINC ION | | Authors: | Massiah, M.A, Matts, J.A.B, Short, K.M, Simmons, B.N, Singireddy, S, Zou, J, Cox, T.C. | | Deposit date: | 2006-05-20 | | Release date: | 2007-04-03 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Solution Structure of the MID1 B-box2 CHC(D/C)C(2)H(2) Zinc-binding Domain: Insights into an Evolutionarily Conserved RING Fold
J.Mol.Biol., 369, 2007
|
|
2JUN
 
 | | Structure of the MID1 tandem B-boxes reveals an interaction reminiscent of intermolecular RING heterodimers | | Descriptor: | Midline-1, ZINC ION | | Authors: | Tao, H, Singireddy, S, Jakkidi, M, Simmons, B.N, Short, K.M, Cox, T.C, Massiah, M.A. | | Deposit date: | 2007-08-31 | | Release date: | 2008-03-04 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Structure of the MID1 Tandem B-Boxes Reveals an Interaction Reminiscent of Intermolecular Ring Heterodimers
Biochemistry, 47, 2008
|
|
3IIA
 
 | | Crystal structure of apo (91-244) RIa subunit of cAMP-dependent protein kinase | | Descriptor: | GLYCEROL, cAMP-dependent protein kinase type I-alpha regulatory subunit | | Authors: | Sjoberg, T.J, Kim, C, Kornev, A.P, Taylor, S.S. | | Deposit date: | 2009-07-31 | | Release date: | 2010-08-11 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Cyclic AMP analog blocks kinase activation by stabilizing inactive conformation: conformational selection highlights a new concept in allosteric inhibitor design.
Mol.Cell Proteomics, 10, 2011
|
|
3PNA
 
 | | Crystal Structure of cAMP bound (91-244)RIa Subunit of cAMP-dependent Protein Kinase | | Descriptor: | ADENOSINE-3',5'-CYCLIC-MONOPHOSPHATE, GLYCEROL, cAMP-dependent protein kinase type I-alpha regulatory subunit | | Authors: | Kim, C, Taylor, S. | | Deposit date: | 2010-11-18 | | Release date: | 2011-02-09 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.503 Å) | | Cite: | Cyclic AMP Analog Blocks Kinase Activation by Stabilizing Inactive Conformation: Conformational Selection Highlights a New Concept in Allosteric Inhibitor Design.
Mol Cell Proteomics, 10, 2011
|
|
4XHV
 
 | | Crystal structure of Drosophila Spinophilin-PDZ and a C-terminal peptide of Neurexin | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, LP20995p, ... | | Authors: | Driller, J.H, Muhammad, K.G.H, Reddy, S, Rey, U, Boehme, M.A, Hollmann, C, Ramesh, N, Depner, H, Luetzkendorf, J, Matkovic, T, Bergeron, D, Quentin, C, Schmoranzer, J, Goettfert, F, Holt, M, Wahl, M.C, Hell, S.W, Walter, A, Sigrist, S.J, Loll, B. | | Deposit date: | 2015-01-06 | | Release date: | 2015-07-01 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.23 Å) | | Cite: | Presynaptic spinophilin tunes neurexin signalling to control active zone architecture and function.
Nat Commun, 6, 2015
|
|
7TRB
 
 | | CRYSTAL STRUCTURE OF FARNESOID X-ACTIVATED RECEPTOR COMPLEXED WITH COMPOUND-32 AKA (1S,3S)-N-({4-[5-(2-FLUOROPR OPAN-2-YL)-1,2,4-OXADIAZOL-3-YL]BICYCLO[2.2.2]OCTAN-1-YL}M ETHYL)-3-HYDROXY-N-[4'-(2-HYDROXYPROPAN-2-YL)-[1,1'-BIPHEN YL]-3-YL]-3-(TRIFLUOROMETHYL)CYCLOBUTANE-1-CARBOXAMIDE | | Descriptor: | (1s,3s)-N-({4-[5-(2-fluoropropan-2-yl)-1,2,4-oxadiazol-3-yl]bicyclo[2.2.2]octan-1-yl}methyl)-3-hydroxy-N-[4'-(2-hydroxypropan-2-yl)[1,1'-biphenyl]-3-yl]-3-(trifluoromethyl)cyclobutane-1-carboxamide, Bile acid receptor, co-activator | | Authors: | Khan, J.A, Ruzanov, M. | | Deposit date: | 2022-01-28 | | Release date: | 2022-06-29 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Discovery of BMS-986339, a Pharmacologically Differentiated Farnesoid X Receptor Agonist for the Treatment of Nonalcoholic Steatohepatitis.
J.Med.Chem., 65, 2022
|
|
7D5L
 
 | | Discovery of BMS-986144, a Third Generation, Pan Genotype NS3/4A Protease Inhibitor for the Treatment of Hepatitis C Virus Infection | | Descriptor: | NS3/4A Protease, ZINC ION, [1,1,1-tris(fluoranyl)-2-methyl-propan-2-yl] ~{N}-[(1~{S},4~{R},6~{S},7~{Z},11~{R},13~{R},14~{S},18~{R})-13-ethyl-18-(7-fluoranyl-6-methoxy-isoquinolin-1-yl)oxy-11-methyl-4-[(1-methylcyclopropyl)sulfonylcarbamoyl]-2,15-bis(oxidanylidene)-3,16-diazatricyclo[14.3.0.0^{4,6}]nonadec-7-en-14-yl]carbamate | | Authors: | Ghosh, K, Anumula, R, Kumar, A. | | Deposit date: | 2020-09-26 | | Release date: | 2020-12-16 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Discovery of BMS-986144, a Third-Generation, Pan-Genotype NS3/4A Protease Inhibitor for the Treatment of Hepatitis C Virus Infection.
J.Med.Chem., 63, 2020
|
|
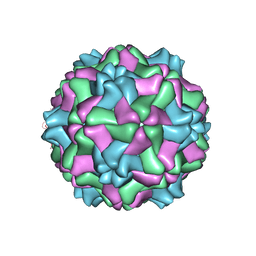
1QJZ
 
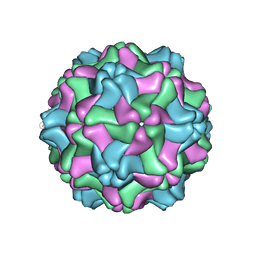 | | Three Dimensional Structure of Physalis Mottle Virus : Implications for the Viral Assembly | | Descriptor: | COAT PROTEIN | | Authors: | Krishna, S.S, Hiremath, C.N, Munshi, S.K, Prahadeeswaran, D, Sastri, M, Savithri, H.S, Murthy, M.R.N. | | Deposit date: | 1999-07-07 | | Release date: | 1999-07-08 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (3.8 Å) | | Cite: | Three Dimensional Structure of Physalis Movirus: Implications for the Viral Assembly
J.Mol.Biol., 289, 1999
|
|
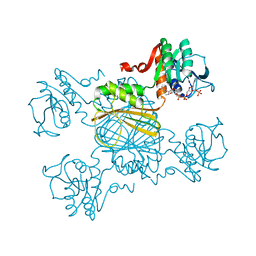
1E57
 
 | |
1DIH
 
 | |
3PA0
 
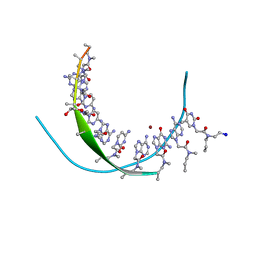 | | Crystal Structure of Chiral Gamma-PNA with Complementary DNA Strand: Insight Into the Stability and Specificity of Recognition an Conformational Preorganization | | Descriptor: | 1,2-ETHANEDIOL, DNA 5'-D(*AP*TP*CP*TP*GP*TP*GP*GP*TP*C)-3', PEPTIDE NUCLEIC ACID, ... | | Authors: | Yeh, J.I, Shivachev, B, Du, S. | | Deposit date: | 2010-10-18 | | Release date: | 2010-12-29 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Crystal structure of chiral gammaPNA with complementary DNA strand: insights into the stability and specificity of recognition and conformational preorganization.
J.Am.Chem.Soc., 132, 2010
|
|
