4LVK
 
 | | MobM Relaxase Domain (MOBV; Mob_Pre) bound to plasmid pMV158 oriT DNA (22nt+3'Phosphate). Mn-bound crystal structure at pH 4.6 | | Descriptor: | ACTTTAT oligonucleotide, ATAAAGTATAGTGTGpo oligonucleotide, MANGANESE (II) ION, ... | | Authors: | Pluta, R, Boer, D.R, Coll, M. | | Deposit date: | 2013-07-26 | | Release date: | 2014-09-24 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.37 Å) | | Cite: | Structural basis of a histidine-DNA nicking/joining mechanism for gene transfer and promiscuous spread of antibiotic resistance.
Proc. Natl. Acad. Sci. U.S.A., 114, 2017
|
|
4LVM
 
 | | MobM Relaxase Domain (MOBV; Mob_Pre) bound to plasmid pMV158 oriT DNA (23nt). Mn-bound crystal structure at pH 6.5 | | Descriptor: | ACTTTAT oligonucleotide, ATAAAGTATAGTGTGT oligonucleotide, CHLORIDE ION, ... | | Authors: | Pluta, R, Boer, D.R, Coll, M. | | Deposit date: | 2013-07-26 | | Release date: | 2014-09-24 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Structural basis of a histidine-DNA nicking/joining mechanism for gene transfer and promiscuous spread of antibiotic resistance.
Proc. Natl. Acad. Sci. U.S.A., 114, 2017
|
|
4LVI
 
 | | MobM Relaxase Domain (MOBV; Mob_Pre) bound to plasmid pMV158 oriT DNA (22nt). Mn-bound crystal structure at pH 4.6 | | Descriptor: | ACTTTAT oligonucleotide, ATAAAGTATAGTGTG oligonucleotide, GLYCEROL, ... | | Authors: | Pluta, R, Boer, D.R, Coll, M. | | Deposit date: | 2013-07-26 | | Release date: | 2014-09-24 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structural basis of a histidine-DNA nicking/joining mechanism for gene transfer and promiscuous spread of antibiotic resistance.
Proc. Natl. Acad. Sci. U.S.A., 114, 2017
|
|
4LVL
 
 | | MobM Relaxase Domain (MOBV; Mob_Pre) bound to plasmid pMV158 oriT DNA (22nt+3'Thiophosphate). Mn-bound crystal structure at pH 6.8 | | Descriptor: | CHLORIDE ION, DNA (5'-D(*AP*CP*TP*TP*TP*AP*T)-3'), DNA (5'-D(*AP*TP*AP*AP*AP*GP*TP*AP*TP*AP*GP*TP*GP*TP*GP*(TS6))-3'), ... | | Authors: | Pluta, R, Boer, D.R, Coll, M. | | Deposit date: | 2013-07-26 | | Release date: | 2014-09-24 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structural basis of a histidine-DNA nicking/joining mechanism for gene transfer and promiscuous spread of antibiotic resistance.
Proc. Natl. Acad. Sci. U.S.A., 114, 2017
|
|
4LVJ
 
 | | MobM Relaxase Domain (MOBV; Mob_Pre) bound to plasmid pMV158 oriT DNA (22nt). Mn-bound crystal structure at pH 5.5 | | Descriptor: | ACETATE ION, ACTTTAT oligonucleotide, ATAAAGTATAGTGTG oligonucleotide, ... | | Authors: | Pluta, R, Boer, D.R, Coll, M. | | Deposit date: | 2013-07-26 | | Release date: | 2014-09-24 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.17 Å) | | Cite: | Structural basis of a histidine-DNA nicking/joining mechanism for gene transfer and promiscuous spread of antibiotic resistance.
Proc. Natl. Acad. Sci. U.S.A., 114, 2017
|
|
7YZ7
 
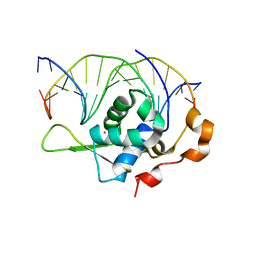 | | Crystal structure of the zebrafish FoxH1 bound to the TGTGGATT site | | Descriptor: | DNA (5'-D(*AP*GP*AP*TP*TP*GP*TP*GP*GP*AP*TP*TP*GP*AP*GP*A)-3'), DNA (5'-D(*TP*CP*TP*CP*AP*AP*TP*CP*CP*AP*CP*AP*AP*TP*CP*T)-3'), Forkhead box protein H1, ... | | Authors: | Pluta, R, Macias, M.J. | | Deposit date: | 2022-02-18 | | Release date: | 2022-11-16 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (0.98 Å) | | Cite: | Molecular basis for DNA recognition by the maternal pioneer transcription factor FoxH1.
Nat Commun, 13, 2022
|
|
7YZB
 
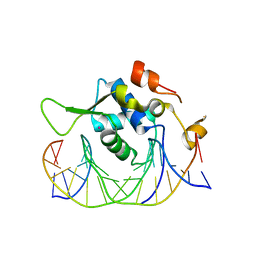 | | Crystal structure of the human FoxH1 bound to the TGTGGATT site | | Descriptor: | DNA (5'-D(*AP*GP*AP*TP*TP*GP*TP*GP*GP*AP*TP*TP*GP*CP*GP*A)-3'), DNA (5'-D(*TP*CP*GP*CP*AP*AP*TP*CP*CP*AP*CP*AP*AP*TP*CP*T)-3'), Forkhead box protein H1, ... | | Authors: | Pluta, R, Macias, M.J. | | Deposit date: | 2022-02-19 | | Release date: | 2022-11-16 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.47 Å) | | Cite: | Molecular basis for DNA recognition by the maternal pioneer transcription factor FoxH1.
Nat Commun, 13, 2022
|
|
7YZD
 
 | |
7YZA
 
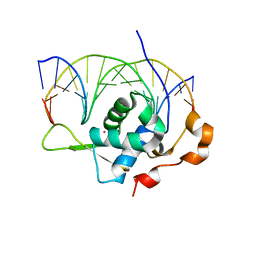 | | Crystal structure of the zebrafish FoxH1 bound to the TGTGTATT site | | Descriptor: | DNA (5'-D(*AP*GP*AP*TP*TP*GP*TP*GP*TP*AP*TP*TP*GP*AP*GP*A)-3'), DNA (5'-D(*TP*CP*TP*CP*AP*AP*TP*AP*CP*AP*CP*AP*AP*TP*CP*T)-3'), Forkhead box protein H1, ... | | Authors: | Pluta, R, Macias, M.J. | | Deposit date: | 2022-02-19 | | Release date: | 2022-11-16 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.18 Å) | | Cite: | Molecular basis for DNA recognition by the maternal pioneer transcription factor FoxH1.
Nat Commun, 13, 2022
|
|
7YZC
 
 | | Crystal structure of the zebrafish FoxH1 bound to the TGTTTATT site | | Descriptor: | ACETATE ION, DNA (5'-D(*AP*GP*AP*TP*TP*GP*TP*TP*TP*AP*TP*TP*GP*AP*GP*A)-3'), DNA (5'-D(*TP*CP*TP*CP*AP*AP*TP*AP*AP*AP*CP*AP*AP*TP*CP*T)-3'), ... | | Authors: | Pluta, R, Macias, M.J. | | Deposit date: | 2022-02-19 | | Release date: | 2022-11-16 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.17 Å) | | Cite: | Molecular basis for DNA recognition by the maternal pioneer transcription factor FoxH1.
Nat Commun, 13, 2022
|
|
7YZG
 
 | | Crystal structure of the Xenopus FoxH1 bound to the TGTGGATT site | | Descriptor: | DNA (5'-D(*CP*AP*GP*AP*TP*TP*GP*TP*GP*GP*AP*TP*TP*GP*AP*G)-3'), DNA (5'-D(*CP*TP*CP*AP*AP*TP*CP*CP*AP*CP*AP*AP*TP*CP*TP*G)-3'), Forkhead box protein H1 | | Authors: | Pluta, R, Macias, M.J. | | Deposit date: | 2022-02-19 | | Release date: | 2022-11-16 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.82 Å) | | Cite: | Molecular basis for DNA recognition by the maternal pioneer transcription factor FoxH1.
Nat Commun, 13, 2022
|
|
7YZF
 
 | |
7YZE
 
 | |
6TBZ
 
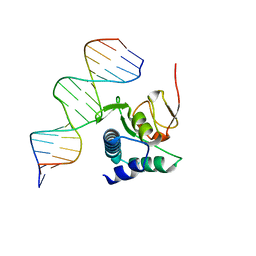 | |
6TCE
 
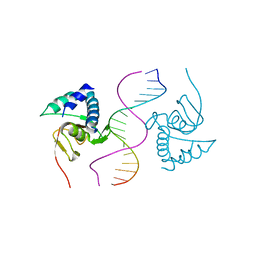 | |
6ZMN
 
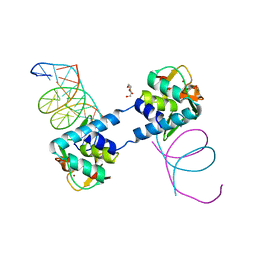 | |
6ZMU
 
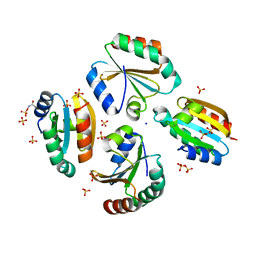 | | Crystal structure of the germline-specific thioredoxin protein Deadhead (Thioredoxin-1) from Drospohila melanogaster, P43212 | | Descriptor: | SODIUM ION, SULFATE ION, Thioredoxin-1 | | Authors: | Baginski, B, Pluta, R, Macias, M.J. | | Deposit date: | 2020-07-03 | | Release date: | 2020-10-21 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Structures of the germline-specific Deadhead and thioredoxin T proteins from Drosophila melanogaster reveal unique features among thioredoxins.
Iucrj, 8, 2021
|
|
6Z7O
 
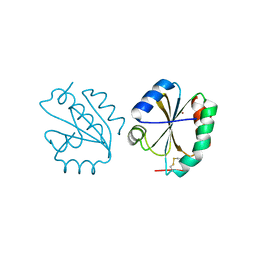 | | Crystal structure of Thioredoxin T from Drosophila melanogaster | | Descriptor: | Thioredoxin-T, ZINC ION | | Authors: | Freier, R, Aragon, E, Baginski, B, Pluta, R, Martin-Malpartida, P, Torner, C, Gonzaez, C, Macias, M. | | Deposit date: | 2020-05-31 | | Release date: | 2020-10-21 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.33 Å) | | Cite: | Structures of the germline-specific Deadhead and thioredoxin T proteins from Drosophila melanogaster reveal unique features among thioredoxins.
Iucrj, 8, 2021
|
|
5N2Q
 
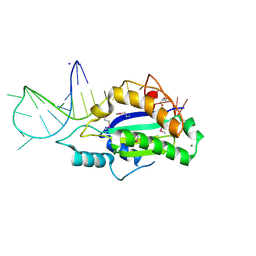 | | MobM Relaxase Domain (MOBV; Mob_Pre) bound to 26nt pMV158 oriT DNA | | Descriptor: | CHLORIDE ION, DNA (26-MER), GLYCEROL, ... | | Authors: | Russi, S, Boer, D.R, Coll, M. | | Deposit date: | 2017-02-08 | | Release date: | 2017-04-12 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural basis of a histidine-DNA nicking/joining mechanism for gene transfer and promiscuous spread of antibiotic resistance.
Proc. Natl. Acad. Sci. U.S.A., 114, 2017
|
|
6FZS
 
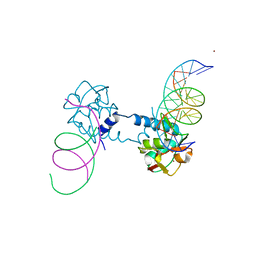 | | Crystal structure of Smad5-MH1 bound to the GGCGC site. | | Descriptor: | DNA (5'-D(P*TP*GP*CP*AP*GP*GP*CP*GP*CP*GP*CP*CP*TP*GP*CP*A)-3'), Mothers against decapentaplegic homolog 5, ZINC ION | | Authors: | Kaczmarska, Z, Marquez, J.A, Macias, M.J. | | Deposit date: | 2018-03-15 | | Release date: | 2019-10-02 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.31 Å) | | Cite: | Unveiling the dimer/monomer propensities of Smad MH1-DNA complexes
Computational and Structural Biotechnology Journal, 19, 2021
|
|
6FZT
 
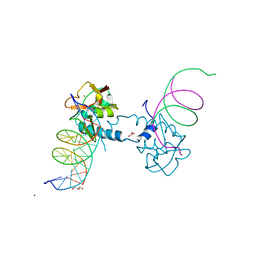 | | Crystal structure of Smad8_9-MH1 bound to the GGCGC site. | | Descriptor: | DNA (5'-D(P*TP*GP*CP*AP*GP*GP*CP*GP*CP*GP*CP*CP*TP*GP*CP*A)-3'), GLYCEROL, Mothers against decapentaplegic homolog 9, ... | | Authors: | Kaczmarska, Z, Marquez, J.A, Macias, M.J. | | Deposit date: | 2018-03-15 | | Release date: | 2019-10-02 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.46 Å) | | Cite: | Unveiling the dimer/monomer propensities of Smad MH1-DNA complexes
Computational and Structural Biotechnology Journal, 19, 2021
|
|
