2YA9
 
 | | Crystal structure of the autoinhibited form of mouse DAPK2 | | Descriptor: | (4S,5S)-1,2-DITHIANE-4,5-DIOL, CALCIUM ION, DEATH-ASSOCIATED PROTEIN KINASE 2 | | Authors: | Patel, A.K, Kursula, P. | | Deposit date: | 2011-02-18 | | Release date: | 2011-04-13 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structure of the Dimeric Autoinhibited Conformation of Dapk2, a Pro-Apoptotic Protein Kinase.
J.Mol.Biol., 409, 2011
|
|
2YAB
 
 | |
2YAA
 
 | |
8CEF
 
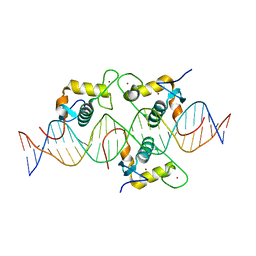 | | Asymmetric Dimerization in a Transcription Factor Superfamily is Promoted by Allosteric Interactions with DNA | | Descriptor: | DNA (26-MER), Nuclear receptor DNA binding domain, ZINC ION | | Authors: | Patel, A.K.M, Shaik, T.B, McEwen, A.G, Moras, D, Klaholz, B.P, Billas, I.M.L. | | Deposit date: | 2023-02-01 | | Release date: | 2023-08-09 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.486 Å) | | Cite: | Asymmetric dimerization in a transcription factor superfamily is promoted by allosteric interactions with DNA.
Nucleic Acids Res., 51, 2023
|
|
6IDN
 
 | | Crystal structure of ICChI chitinase from ipomoea carnea | | Descriptor: | CALCIUM ION, ICChI, a glycosylated chitinase, ... | | Authors: | Kumar, S, Kumar, A, Patel, A.K. | | Deposit date: | 2018-09-10 | | Release date: | 2018-11-21 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | TIM barrel fold and glycan moieties in the structure of ICChI, a protein with chitinase and lysozyme activity.
Phytochemistry, 170, 2020
|
|
8HMS
 
 | | Crystal Structure of PKM2 mutant C474S | | Descriptor: | 1,2-ETHANEDIOL, 1,6-di-O-phosphono-beta-D-fructofuranose, GLYCEROL, ... | | Authors: | Upadhyay, S, Kumar, A, Patel, A.K. | | Deposit date: | 2022-12-05 | | Release date: | 2023-01-11 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structural and mechanistic insights into cancer patient-derived mutations in Pyruvate Kinase muscle isoform 2
To Be Published
|
|
8HMQ
 
 | | Crystal Structure of PKM2 mutant P403A | | Descriptor: | 1,2-ETHANEDIOL, 1,6-di-O-phosphono-beta-D-fructofuranose, GLYCEROL, ... | | Authors: | Upadhyay, S, Kumar, A, Patel, A.K. | | Deposit date: | 2022-12-05 | | Release date: | 2023-01-11 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structural and mechanistic insights into cancer patient-derived mutations in Pyruvate Kinase muscle isoform 2
To Be Published
|
|
8HMR
 
 | | Crystal Structure of PKM2 mutant L144P | | Descriptor: | 1,2-ETHANEDIOL, 1,6-di-O-phosphono-beta-D-fructofuranose, MAGNESIUM ION, ... | | Authors: | Upadhyay, S, Kumar, A, Patel, A.K. | | Deposit date: | 2022-12-05 | | Release date: | 2023-01-11 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structural and mechanistic insights into cancer patient-derived mutations in Pyruvate Kinase muscle isoform 2
To Be Published
|
|
8HMU
 
 | | Crystal Structure of PKM2 mutant R516C | | Descriptor: | 1,2-ETHANEDIOL, 1,6-di-O-phosphono-beta-D-fructofuranose, GLYCEROL, ... | | Authors: | Upadhyay, S, Kumar, A, Patel, A.K. | | Deposit date: | 2022-12-05 | | Release date: | 2023-01-11 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structural and mechanistic insights into cancer patient-derived mutations in Pyruvate Kinase muscle isoform 2
To Be Published
|
|
8AAG
 
 | | H1-bound palindromic nucleosome, state 1 | | Descriptor: | DNA/RNA (185-MER), Histone H1.0-B, Histone H2A type 1, ... | | Authors: | Alegrio Louro, J, Beinsteiner, B, Cheng, T.C, Patel, A.K.M, Boopathi, R, Angelov, D, Hamiche, A, Bednar, J, Kale, S, Dimitrov, S, Klaholz, B. | | Deposit date: | 2022-07-01 | | Release date: | 2022-12-14 | | Last modified: | 2024-07-24 | | Method: | ELECTRON MICROSCOPY (10 Å) | | Cite: | Nucleosome dyad determines the H1 C-terminus collapse on distinct DNA arms.
Structure, 31, 2023
|
|
6JHU
 
 | | Crystal Structure Of Biotin Protein Ligase From Leishmania Major in complex with Biotinyl-5-AMP | | Descriptor: | BIOTINYL-5-AMP, Biotin/lipoate protein ligase-like protein, SULFATE ION | | Authors: | Rajak, M, Patel, A, Sundd, M. | | Deposit date: | 2019-02-19 | | Release date: | 2020-04-08 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.97 Å) | | Cite: | Leishmania major biotin protein ligase forms a unique cross-handshake dimer
Acta Crystallogr.,Sect.D, 77, 2021
|
|
7DBS
 
 | |
5ZWS
 
 | |
5ZWT
 
 | |
7Q71
 
 | | The crystallographic structure of the Ligand Binding domain of the NR7 nuclear receptor from the amphioxus Branchiostoma lanceolatum | | Descriptor: | CHLORIDE ION, Nuclear hormone receptor 7, PHOSPHATE ION | | Authors: | Billas, I.M.L, McEwen, A.G, Hazemann, I, Moras, D, Laudet, V. | | Deposit date: | 2021-11-09 | | Release date: | 2022-09-14 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | A novel nuclear receptor subfamily enlightens the origin of heterodimerization.
Bmc Biol., 20, 2022
|
|
