1GP8
 
 | | NMR SOLUTION STRUCTURE OF THE COAT PROTEIN-BINDING DOMAIN OF BACTERIOPHAGE P22 SCAFFOLDING PROTEIN | | Descriptor: | PROTEIN (SCAFFOLDING PROTEIN) | | Authors: | Sun, Y, Parker, M.H, Weigele, P, Casjens, S, Prevelige Jr, P.E, Krishna, N.R. | | Deposit date: | 1999-05-11 | | Release date: | 1999-05-17 | | Last modified: | 2023-12-27 | | Method: | SOLUTION NMR | | Cite: | Structure of the coat protein-binding domain of the scaffolding protein from a double-stranded DNA virus.
J.Mol.Biol., 297, 2000
|
|
2GP8
 
 | | NMR SOLUTION STRUCTURE OF THE COAT PROTEIN-BINDING DOMAIN OF BACTERIOPHAGE P22 SCAFFOLDING PROTEIN | | Descriptor: | PROTEIN (SCAFFOLDING PROTEIN) | | Authors: | Sun, Y, Parker, M.H, Weigele, P, Casjens, S, Prevelige Jr, P.E, Krishna, N.R. | | Deposit date: | 1999-05-11 | | Release date: | 1999-05-17 | | Last modified: | 2023-12-27 | | Method: | SOLUTION NMR | | Cite: | Structure of the coat protein-binding domain of the scaffolding protein from a double-stranded DNA virus.
J.Mol.Biol., 297, 2000
|
|
1EKU
 
 | | CRYSTAL STRUCTURE OF A BIOLOGICALLY ACTIVE SINGLE CHAIN MUTANT OF HUMAN IFN-GAMMA | | Descriptor: | Interferon gamma, SULFATE ION | | Authors: | Landar, A, Curry, B, Parker, M.H, DiGiacomo, R, Indelicato, S.R, Walter, M.R. | | Deposit date: | 2000-03-09 | | Release date: | 2000-09-01 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Design, characterization, and structure of a biologically active single-chain mutant of human IFN-gamma.
J.Mol.Biol., 299, 2000
|
|
2Q11
 
 | | Structure of BACE complexed to compound 1 | | Descriptor: | 3-(2-AMINO-6-BENZOYLQUINAZOLIN-3(4H)-YL)-N-CYCLOHEXYL-N-METHYLPROPANAMIDE, Beta-secretase 1 | | Authors: | Sharff, A.J. | | Deposit date: | 2007-05-23 | | Release date: | 2007-08-14 | | Last modified: | 2021-10-20 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | 2-Amino-3,4-dihydroquinazolines as inhibitors of BACE-1 (beta-Site APP cleaving enzyme): Use of structure based design to convert a micromolar hit into a nanomolar lead.
J.Med.Chem., 50, 2007
|
|
2Q15
 
 | | Structure of BACE complexed to compound 3a | | Descriptor: | (4S)-4-(2-AMINO-6-PHENOXYQUINAZOLIN-3(4H)-YL)-N,4-DICYCLOHEXYL-N-METHYLBUTANAMIDE, Beta-secretase 1 | | Authors: | Sharff, A.J. | | Deposit date: | 2007-05-23 | | Release date: | 2007-08-14 | | Last modified: | 2021-10-20 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | 2-Amino-3,4-dihydroquinazolines as inhibitors of BACE-1 (beta-Site APP cleaving enzyme): Use of structure based design to convert a micromolar hit into a nanomolar lead.
J.Med.Chem., 50, 2007
|
|
5C37
 
 | | Structure of the beta-ketoacyl reductase domain of human fatty acid synthase bound to a spiro-imidazolone inhibitor | | Descriptor: | 6-{[(3R)-1-(cyclopropylcarbonyl)pyrrolidin-3-yl]methyl}-5-[4-(1-methyl-1H-indazol-5-yl)phenyl]-4,6-diazaspiro[2.4]hept-4-en-7-one, CHLORIDE ION, Fatty acid synthase, ... | | Authors: | Schubert, C, Milligan, C.M, Vo, K, Grasberger, B. | | Deposit date: | 2015-06-17 | | Release date: | 2016-06-22 | | Last modified: | 2024-06-19 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Design and synthesis of a series of bioavailable fatty acid synthase (FASN) KR domain inhibitors for cancer therapy.
Bioorg.Med.Chem.Lett., 28, 2018
|
|
2JC2
 
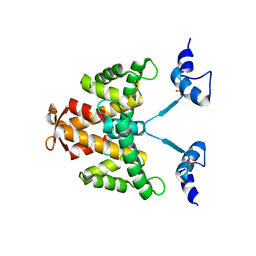 | | The crystal structure of the natural F112L human sorcin mutant | | Descriptor: | SORCIN, SULFATE ION | | Authors: | Franceschini, S, Ilari, A, Colotti, G, Chiancone, E. | | Deposit date: | 2006-12-19 | | Release date: | 2007-08-28 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Molecular Basis for the Impaired Function of the Natural F112L Sorcin Mutant: X-Ray Crystal Structure, Calcium Affinity, and Interaction with Annexin Vii and the Ryanodine Receptor.
Faseb J., 22, 2008
|
|
1JUO
 
 | |
1LK3
 
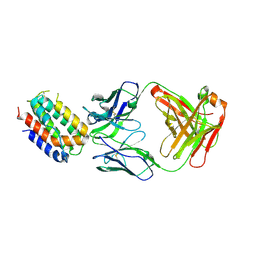 | | ENGINEERED HUMAN INTERLEUKIN-10 MONOMER COMPLEXED TO 9D7 FAB FRAGMENT | | Descriptor: | 9D7 Heavy Chain, 9D7 Light Chain, Interleukin-10 | | Authors: | Josephson, K, Jones, B.C, Walter, L.J, DiGiacomo, R, Indelicato, S.R, Walter, M.R. | | Deposit date: | 2002-04-23 | | Release date: | 2002-07-17 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.91 Å) | | Cite: | Noncompetitive antibody neutralization of IL-10 revealed by protein engineering and x-ray crystallography.
Structure, 10, 2002
|
|
