2DS6
 
 | | Structure of the ZBD in the tetragonal crystal form | | Descriptor: | ATP-dependent Clp protease ATP-binding subunit clpX, ZINC ION | | Authors: | Park, E.Y, Lee, B.G, Hong, S.B, Song, H.K. | | Deposit date: | 2006-06-22 | | Release date: | 2007-02-13 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural Basis of SspB-tail Recognition by the Zinc Binding Domain of ClpX.
J.Mol.Biol., 367, 2007
|
|
2DS8
 
 | | Structure of the ZBD-XB complex | | Descriptor: | ATP-dependent Clp protease ATP-binding subunit clpX, SspB-tail peptide, ZINC ION | | Authors: | Park, E.Y, Lee, B.G, Hong, S.B, Kim, H.W, Song, H.K. | | Deposit date: | 2006-06-22 | | Release date: | 2007-02-13 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Structural Basis of SspB-tail Recognition by the Zinc Binding Domain of ClpX.
J.Mol.Biol., 367, 2007
|
|
2DS7
 
 | | Structure of the ZBD in the hexagonal crystal form | | Descriptor: | ATP-dependent Clp protease ATP-binding subunit clpX, ZINC ION | | Authors: | Park, E.Y, Lee, B.G, Hong, S.B, Song, H.K. | | Deposit date: | 2006-06-22 | | Release date: | 2007-02-13 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structural Basis of SspB-tail Recognition by the Zinc Binding Domain of ClpX.
J.Mol.Biol., 367, 2007
|
|
3KB5
 
 | |
2H8G
 
 | | 5'-Methylthioadenosine Nucleosidase from Arabidopsis thaliana | | Descriptor: | 5'-Methylthioadenosine Nucleosidase, ADENINE | | Authors: | Park, E.Y, Oh, S.I, Nam, M.J, Shin, J.S, Kim, K.N, Song, H.K. | | Deposit date: | 2006-06-07 | | Release date: | 2006-10-10 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Crystal structure of 5'-methylthioadenosine nucleosidase from Arabidopsis thaliana at 1.5-A resolution
Proteins, 65, 2006
|
|
1TX6
 
 | | trypsin:BBI complex | | Descriptor: | Bowman-Birk type trypsin inhibitor, CALCIUM ION, Trypsin | | Authors: | Song, H.K, Park, E.Y, Kim, J.A, Kim, H.W, Kim, Y.S. | | Deposit date: | 2004-07-02 | | Release date: | 2005-03-08 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal structure of the Bowman-Birk inhibitor from barley seeds in ternary complex with porcine trypsin
J.Mol.Biol., 343, 2004
|
|
2DS5
 
 | | Structure of the ZBD in the orthorhomibic crystal from | | Descriptor: | ATP-dependent Clp protease ATP-binding subunit clpX, CALCIUM ION, TETRAETHYLENE GLYCOL, ... | | Authors: | Song, H.K, Park, E.Y, Lee, B.G, Hong, S.B. | | Deposit date: | 2006-06-22 | | Release date: | 2007-02-13 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Structural Basis of SspB-tail Recognition by the Zinc Binding Domain of ClpX.
J.Mol.Biol., 367, 2007
|
|
3BSF
 
 | |
7WLG
 
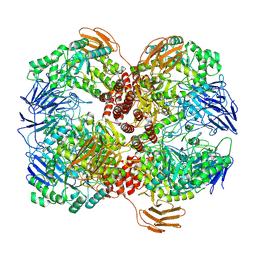 | | Cryo-EM structure of GH31 alpha-1,3-glucosidase from Lactococcus lactis subsp. cremoris | | Descriptor: | Alpha-xylosidase | | Authors: | Ikegaya, M, Moriya, T, Adachi, N, Kawasaki, M, Park, E.Y, Miyazaki, T. | | Deposit date: | 2022-01-13 | | Release date: | 2022-03-30 | | Last modified: | 2024-06-26 | | Method: | ELECTRON MICROSCOPY (2.73 Å) | | Cite: | Structural basis of the strict specificity of a bacterial GH31 alpha-1,3-glucosidase for nigerooligosaccharides.
J.Biol.Chem., 298, 2022
|
|
1R6M
 
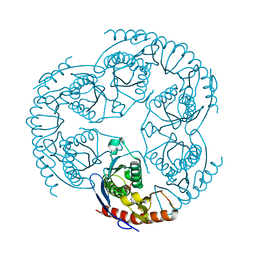 | | Crystal Structure Of The tRNA Processing Enzyme Rnase pH From Pseudomonas Aeruginosa In Complex With Phosphate | | Descriptor: | PHOSPHATE ION, Ribonuclease PH | | Authors: | Choi, J.M, Park, E.Y, Kim, J.H, Chang, S.K, Cho, Y. | | Deposit date: | 2003-10-15 | | Release date: | 2004-02-17 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Probing the functional importance of the hexameric ring structure of RNase PH
J.BIOL.CHEM., 279, 2004
|
|
1R6L
 
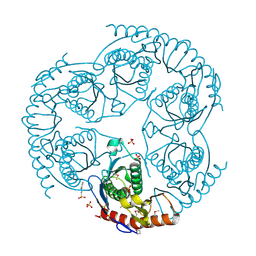 | | Crystal Structure Of The tRNA Processing Enzyme Rnase pH From Pseudomonas Aeruginosa | | Descriptor: | 2-[N-CYCLOHEXYLAMINO]ETHANE SULFONIC ACID, Ribonuclease PH, SULFATE ION | | Authors: | Choi, J.M, Park, E.Y, Kim, J.H, Chang, S.K, Cho, Y. | | Deposit date: | 2003-10-15 | | Release date: | 2004-02-17 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Probing the functional importance of the hexameric ring structure of RNase PH
J.BIOL.CHEM., 279, 2004
|
|
6LGE
 
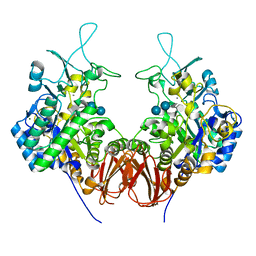 | | Bombyx mori GH13 sucrose hydrolase complexed with acarbose | | Descriptor: | 4,6-dideoxy-4-{[(1S,4R,5S,6S)-4,5,6-trihydroxy-3-(hydroxymethyl)cyclohex-2-en-1-yl]amino}-alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose-(1-4)-beta-D-glucopyranose, CALCIUM ION, MAGNESIUM ION, ... | | Authors: | Miyazaki, T. | | Deposit date: | 2019-12-05 | | Release date: | 2020-05-20 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Structure-function analysis of silkworm sucrose hydrolase uncovers the mechanism of substrate specificity in GH13 subfamily 17exo-alpha-glucosidases.
J.Biol.Chem., 295, 2020
|
|
6LGF
 
 | | Bombyx mori GH13 sucrose hydrolase mutant D247N complexed with sucrose | | Descriptor: | CALCIUM ION, MAGNESIUM ION, Sucrose hydrolase, ... | | Authors: | Miyazaki, T. | | Deposit date: | 2019-12-05 | | Release date: | 2020-05-20 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Structure-function analysis of silkworm sucrose hydrolase uncovers the mechanism of substrate specificity in GH13 subfamily 17exo-alpha-glucosidases.
J.Biol.Chem., 295, 2020
|
|
6LGB
 
 | | Bombyx mori GH13 sucrose hydrolase complexed with glucose | | Descriptor: | CALCIUM ION, GLYCEROL, MAGNESIUM ION, ... | | Authors: | Miyazaki, T. | | Deposit date: | 2019-12-05 | | Release date: | 2020-05-20 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structure-function analysis of silkworm sucrose hydrolase uncovers the mechanism of substrate specificity in GH13 subfamily 17exo-alpha-glucosidases.
J.Biol.Chem., 295, 2020
|
|
6M76
 
 | | GH31 alpha-N-acetylgalactosaminidase from Enterococcus faecalis | | Descriptor: | 1,2-ETHANEDIOL, LPXTG-motif cell wall anchor domain protein | | Authors: | Miyazaki, T. | | Deposit date: | 2020-03-17 | | Release date: | 2020-05-20 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Crystal structure of the Enterococcus faecalis alpha-N-acetylgalactosaminidase, a member of the glycoside hydrolase family 31.
Febs Lett., 594, 2020
|
|
6M77
 
 | |
7BWB
 
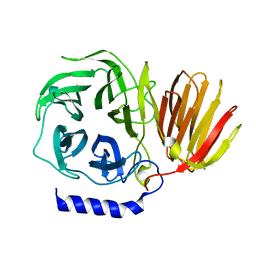 | | Bombyx mori GH32 beta-fructofuranosidase BmSUC1 | | Descriptor: | Beta-fructofuranosidase | | Authors: | Miyazaki, T, Oba, N. | | Deposit date: | 2020-04-14 | | Release date: | 2020-11-04 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structural insight into the substrate specificity of Bombyx mori beta-fructofuranosidase belonging to the glycoside hydrolase family 32.
Insect Biochem.Mol.Biol., 127, 2020
|
|
7BWC
 
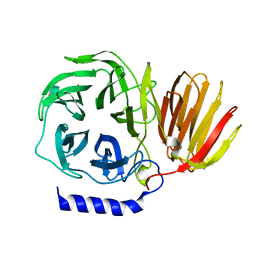 | |
3WO4
 
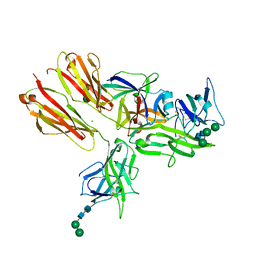 | | Crystal structure of the IL-18 signaling ternary complex | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Tsutsumi, N, Kimura, T, Arita, K, Ariyoshi, M, Ohnishi, H, Kondo, N, Shirakawa, M, Kato, Z, Tochio, H. | | Deposit date: | 2013-12-19 | | Release date: | 2014-12-17 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | The structural basis for receptor recognition of human interleukin-18
Nat Commun, 5, 2014
|
|
3WO2
 
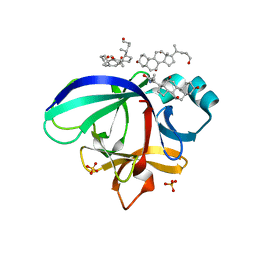 | | Crystal structure of human interleukin-18 | | Descriptor: | 3-[(3-CHOLAMIDOPROPYL)DIMETHYLAMMONIO]-1-PROPANESULFONATE, Interleukin-18, SULFATE ION | | Authors: | Tsutsumi, N, Kimura, T, Arita, K, Ariyoshi, M, Ohnishi, H, Kondo, N, Shirakawa, M, Kato, Z, Tochio, H. | | Deposit date: | 2013-12-19 | | Release date: | 2014-12-17 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.33 Å) | | Cite: | The structural basis for receptor recognition of human interleukin-18
Nat Commun, 5, 2014
|
|
3WO3
 
 | | Crystal structure of IL-18 in complex with IL-18 receptor alpha | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Tsutsumi, N, Kimura, T, Arita, K, Ariyoshi, M, Ohnishi, H, Kondo, N, Shirakawa, M, Kato, Z, Tochio, H. | | Deposit date: | 2013-12-19 | | Release date: | 2014-12-17 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | The structural basis for receptor recognition of human interleukin-18
Nat Commun, 5, 2014
|
|
8IUA
 
 | |
8IU9
 
 | |
8IU8
 
 | |
8IUB
 
 | |
