3KDR
 
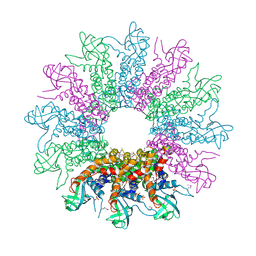 | | The Crystal Structure of a HK97 Family Phage Portal Protein from Corynebacterium diphtheriae to 2.9A | | Descriptor: | GLYCEROL, HK97 Family Phage Portal Protein, PHOSPHATE ION, ... | | Authors: | Nocek, B, Stein, A.J, Mulligan, R, Duggan, E, Abdullah, J, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2009-10-23 | | Release date: | 2009-12-29 | | Last modified: | 2017-11-01 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | The Crystal Structure of a HK97 Family Phage Portal Protein from Corynebacterium diphtheriae to 2.9A
To be Published
|
|
3C2Q
 
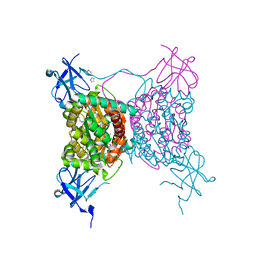 | | Crystal structure of conserved putative LOR/SDH protein from Methanococcus maripaludis S2 | | Descriptor: | IMIDAZOLE, NICKEL (II) ION, Uncharacterized conserved protein | | Authors: | Duke, N, Gu, M, Mulligan, R, Conrad, B, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2008-01-25 | | Release date: | 2008-02-05 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structure of conserved putative LOR/SDH protein from Methanococcus maripaludis S2
To be Published
|
|
4X3Z
 
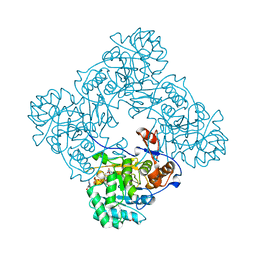 | | Inosine 5'-monophosphate dehydrogenase from Vibrio cholerae, deletion mutant, in complex with XMP and NAD | | Descriptor: | GLYCEROL, Inosine-5'-monophosphate dehydrogenase, NICOTINAMIDE-ADENINE-DINUCLEOTIDE, ... | | Authors: | Osipiuk, J, MALTSEVA, N, KIM, Y, Mulligan, R, MAKOWSKA-GRZYSKA, M, Gu, M, Anderson, W.F, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2014-12-02 | | Release date: | 2014-12-10 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.62 Å) | | Cite: | Inosine 5'-monophosphate dehydrogenase from Vibrio cholerae, deletion mutant, in complex with XMP and NAD
to be published
|
|
4RN7
 
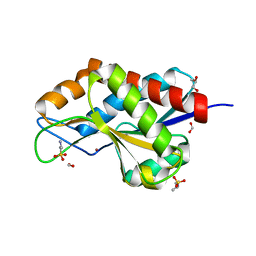 | | The crystal structure of N-acetylmuramoyl-L-alanine amidase from Clostridium difficile 630 | | Descriptor: | 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, FORMIC ACID, GLYCEROL, ... | | Authors: | Tan, K, Mulligan, R, Kwon, K, Anderson, W.F, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2014-10-23 | | Release date: | 2014-11-05 | | Last modified: | 2017-11-22 | | Method: | X-RAY DIFFRACTION (1.717 Å) | | Cite: | The crystal structure of N-acetylmuramoyl-L-alanine amidase from Clostridium difficile 630
To be Published
|
|
3IIE
 
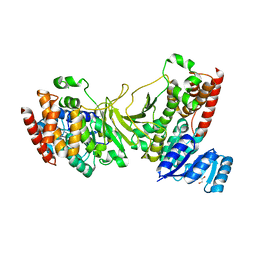 | | 1-deoxy-D-xylulose 5-phosphate reductoisomerase from Yersinia pestis. | | Descriptor: | 1,2-ETHANEDIOL, 1-deoxy-D-xylulose 5-phosphate reductoisomerase, MAGNESIUM ION | | Authors: | Osipiuk, J, Mulligan, R, Stam, J, Anderson, W.F, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2009-07-31 | | Release date: | 2009-08-11 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.21 Å) | | Cite: | X-ray crystal structure of 1-deoxy-D-xylulose 5-phosphate reductoisomerase from Yersinia pestis.
To be Published
|
|
6PU9
 
 | | Crystal Structure of the Type B Chloramphenicol O-Acetyltransferase from Vibrio vulnificus | | Descriptor: | 1,2-ETHANEDIOL, Acetyltransferase, CHLORIDE ION | | Authors: | Kim, Y, Maltseva, N, Mulligan, R, Grimshaw, S, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2019-07-17 | | Release date: | 2019-08-14 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structural and functional characterization of three Type B and C chloramphenicol acetyltransferases from Vibrio species.
Protein Sci., 29, 2020
|
|
3BIO
 
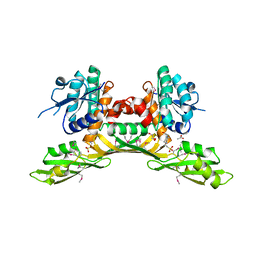 | | Crystal structure of oxidoreductase (Gfo/Idh/MocA family member) from Porphyromonas gingivalis W83 | | Descriptor: | 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, ACETATE ION, GLYCEROL, ... | | Authors: | Nocek, B, Mulligan, R, Moy, S, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2007-11-30 | | Release date: | 2007-12-25 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal structure of oxidoreductase (Gfo/Idh/MocA family member) from Porphyromonas gingivalis W83.
To be Published
|
|
3B8B
 
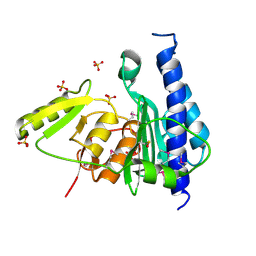 | | Crystal structure of CysQ from Bacteroides thetaiotaomicron, a bacterial member of the inositol monophosphatase family | | Descriptor: | CHLORIDE ION, CysQ, sulfite synthesis pathway protein, ... | | Authors: | Cuff, M.E, Mulligan, R, Abdullah, J, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2007-10-31 | | Release date: | 2007-12-18 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | The structure of CysQ from Bacteroides thetaiotaomicron, a bacterial member of the inositol monophosphatase family.
TO BE PUBLISHED
|
|
7TCB
 
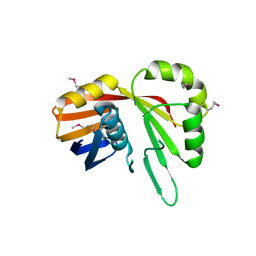 | | Crystal Structure of the YaeQ Family Protein VPA0551 from Vibrio parahaemolyticus | | Descriptor: | YaeQ family protein VPA0551 | | Authors: | Kim, Y, Mulligan, R, Maltseva, N, Gu, M, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2021-12-23 | | Release date: | 2022-01-05 | | Last modified: | 2023-04-19 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Crystal Structure of the YaeQ Family Protein VPA0551 from Vibrio parahaemolyticus
To Be Published
|
|
5HI6
 
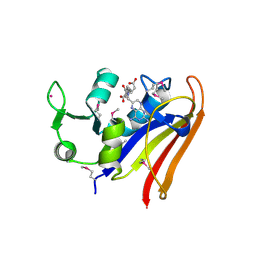 | | The high resolution structure of dihydrofolate reductase from Yersinia pestis complex with methotrexate as closed form | | Descriptor: | CALCIUM ION, CHLORIDE ION, Dihydrofolate reductase, ... | | Authors: | Chang, C, Maltseva, N, Kim, Y, Makowska-Grzyska, M, Mulligan, R, Papazisi, L, Anderson, W.F, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2016-01-11 | | Release date: | 2016-02-03 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.051 Å) | | Cite: | The high resolution structure of dihydrofolate reductase from Yersinia pestis complex with methotrexate as closed form
To Be Published
|
|
5HD6
 
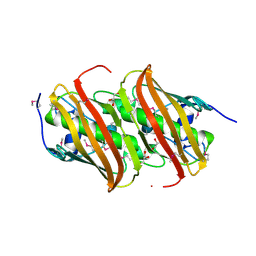 | | High resolution structure of 3-hydroxydecanoyl-(acyl carrier protein) dehydratase from Yersinia pestis at 1.35 A | | Descriptor: | 3-hydroxydecanoyl-[acyl-carrier-protein] dehydratase, GLYCEROL | | Authors: | Chang, C, Maltseva, N, Kim, Y, Mulligan, R, Kwon, K, Anderson, W.F, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2016-01-04 | | Release date: | 2016-01-20 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (1.35 Å) | | Cite: | High resolution structure of 3-hydroxydecanoyl-(acyl carrier protein) dehydratase from Yersinia pestis at 1.35 A
To Be Published
|
|
5FDA
 
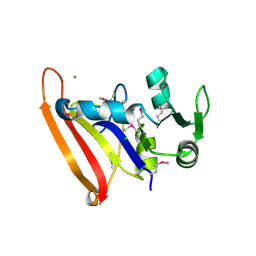 | | The high resolution structure of apo form dihydrofolate reductase from Yersinia pestis at 1.55 A | | Descriptor: | CHLORIDE ION, Dihydrofolate reductase | | Authors: | Chang, C, Maltseva, N, Kim, Y, Makowska-Grzyska, M, Mulligan, R, Papazisi, L, Anderson, W.F, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2015-12-15 | | Release date: | 2015-12-30 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.549 Å) | | Cite: | structure of dihydrofolate reductase from Yersinia pestis complex with
To Be Published
|
|
5DUL
 
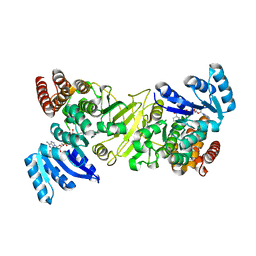 | | 1-deoxy-D-xylulose 5-phosphate reductoisomerase from Yersinia pestis in complex with NADPH | | Descriptor: | 1-deoxy-D-xylulose 5-phosphate reductoisomerase, NADPH DIHYDRO-NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE | | Authors: | Osipiuk, J, Mulligan, R, Stam, J, Anderson, W.F, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2015-09-18 | | Release date: | 2015-09-30 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | 1-deoxy-D-xylulose 5-phosphate reductoisomerase from Yersinia pestis in complex with NADPH .
to be published
|
|
5F4C
 
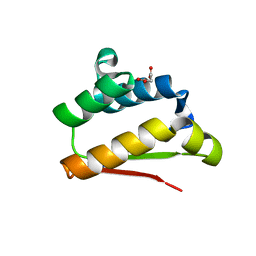 | | Crystal Structure of Ribonuclease Inhibitor Barstar from Salmonella Typhimurium | | Descriptor: | MALONATE ION, Putative cytoplasmic protein | | Authors: | Maltseva, N, Kim, Y, Mulligan, R, Stam, J, Anderson, W.F, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2015-12-03 | | Release date: | 2015-12-23 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Crystal Structure of Ribonuclease Inhibitor Barstar from Salmonella Typhimurium.
To Be Published
|
|
5F64
 
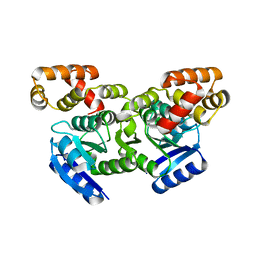 | | Putative positive transcription regulator (sensor EvgS) from Shigella flexneri | | Descriptor: | Positive transcription regulator EvgA | | Authors: | Nocek, B, Osipiuk, J, Mulligan, R, Gu, M, Anderson, W.F, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2015-12-05 | | Release date: | 2015-12-30 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.71 Å) | | Cite: | Putative positive transcription regulator (sensor EvgS) from Shigella flexneri.
to be published
|
|
5EYF
 
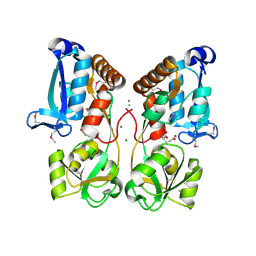 | | Crystal Structure of Solute-binding Protein from Enterococcus faecium with Bound Glutamate | | Descriptor: | CHLORIDE ION, GLUTAMIC ACID, Glutamate ABC superfamily ATP binding cassette transporter, ... | | Authors: | Maltseva, N, Kim, Y, Mulligan, R, Shatsman, S, Anderson, W.F, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2015-11-24 | | Release date: | 2015-12-16 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.52 Å) | | Cite: | Crystal Structure of Solute-binding Protein from Enterococcus faecium with Bound Glutamate
To Be Published
|
|
2QSA
 
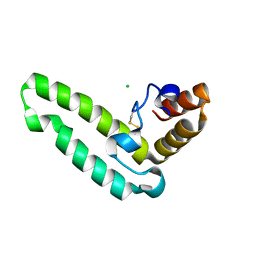 | | Crystal structure of J-domain of DnaJ homolog dnj-2 precursor from C.elegans. | | Descriptor: | CHLORIDE ION, DnaJ homolog dnj-2 | | Authors: | Osipiuk, J, Mulligan, R, Gu, M, Voisine, C, Morimoto, R.I, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2007-07-30 | | Release date: | 2007-08-14 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (1.68 Å) | | Cite: | X-ray crystal structure of J-domain of DnaJ homolog dnj-2 precursor from C.elegans.
To be Published
|
|
2R6H
 
 | | Crystal structure of the domain comprising the NAD binding and the FAD binding regions of the NADH:ubiquinone oxidoreductase, Na translocating, F subunit from Porphyromonas gingivalis | | Descriptor: | FLAVIN-ADENINE DINUCLEOTIDE, NADH:ubiquinone oxidoreductase, Na translocating, ... | | Authors: | Kim, Y, Mulligan, R, Moy, S, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2007-09-05 | | Release date: | 2007-09-11 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.95 Å) | | Cite: | Crystal Structure of the Domain Comprising the Regions Binding NAD and FAD from the NADH:Ubiquinone Oxidoreductase, Na Translocating, F Subunit from Porphyromonas gingivalis.
To be Published
|
|
2R8R
 
 | | Crystal structure of the N-terminal region (19..243) of sensor protein KdpD from Pseudomonas syringae pv. tomato str. DC3000 | | Descriptor: | DI(HYDROXYETHYL)ETHER, GLYCEROL, SULFATE ION, ... | | Authors: | Nocek, B, Mulligan, R, Moy, S, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2007-09-11 | | Release date: | 2007-09-25 | | Last modified: | 2017-10-25 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal structure of the N-terminal region (19..243) of sensor protein KdpD from Pseudomonas syringae pv. tomato str. DC3000.
To be Published
|
|
2AU5
 
 | |
2F7W
 
 | |
2F7Y
 
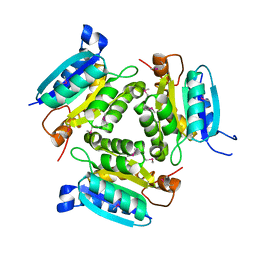 | |
2FB6
 
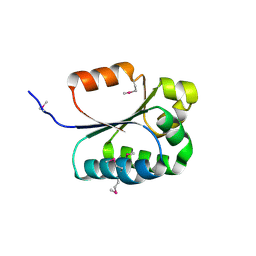 | | Structure of Conserved Protein of Unknown Function BT1422 from Bacteroides thetaiotaomicron | | Descriptor: | conserved hypothetical protein | | Authors: | Osipiuk, J, Mulligan, R, Abdullah, J, Collart, F, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2005-12-08 | | Release date: | 2006-01-24 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.46 Å) | | Cite: | X-ray crystal structure of conserved hypothetical protein BT1422 from Bacteroides thetaiotaomicron
To be Published
|
|
2HSJ
 
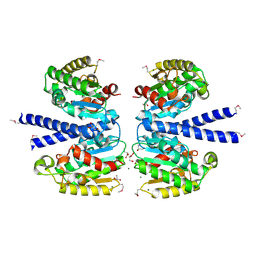 | | The structure of a putative platelet activating factor from Streptococcus pneumonia. | | Descriptor: | GLYCEROL, MAGNESIUM ION, Putative platelet activating factor | | Authors: | Cuff, M.E, Mulligan, R, Abdullah, J, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2006-07-21 | | Release date: | 2006-09-19 | | Last modified: | 2017-10-18 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | The structure of a putative platelet activating factor from Streptococcus pneumonia.
To be Published
|
|
2IAF
 
 | |
