3DG9
 
 | | Crystal Structure of Malonate Decarboxylase from Bordatella bronchiseptica | | Descriptor: | Arylmalonate decarboxylase, PHOSPHATE ION | | Authors: | Okrasa, K, Levy, C, Baudendistel, N, Leys, D, Micklefield, J. | | Deposit date: | 2008-06-13 | | Release date: | 2008-08-05 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Structure and Mechanism of an Unusual Malonate Decarboxylase and Related Racemases.
Chemistry, 14, 2008
|
|
1XT7
 
 | | Daptomycin NMR Structure | | Descriptor: | DAPTOMYCIN, DECANOIC ACID | | Authors: | Ball, L.-J, Goult, C.M, Donarski, J.A, Micklefield, J, Ramesh, V. | | Deposit date: | 2004-10-21 | | Release date: | 2004-11-16 | | Last modified: | 2012-12-12 | | Method: | SOLUTION NMR | | Cite: | NMR Structure Determination and Calcium Binding Effects of Lipopeptide Antibiotic Daptomycin
Org.Biomol.Chem., 2, 2004
|
|
4Z44
 
 | | F454K Mutant of Tryptophan 7-halogenase PrnA | | Descriptor: | CHLORIDE ION, FLAVIN-ADENINE DINUCLEOTIDE, Flavin-dependent tryptophan halogenase PrnA, ... | | Authors: | Shepherd, S.A, Karthikeyan, C, Latham, J, Struck, A.-W, Thompson, M.L, Menon, B, Levy, C.W, Leys, D, Micklefield, J. | | Deposit date: | 2015-04-01 | | Release date: | 2016-04-13 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.204 Å) | | Cite: | Extending the biocatalytic scope of regiocomplementary flavin-dependent halogenase enzymes.
Chem Sci, 6, 2015
|
|
3IP8
 
 | | Crystal structure of arylmalonate decarboxylase (AMDase) from Bordatella bronchiseptic in complex with benzylphosphonate | | Descriptor: | Arylmalonate decarboxylase, benzylphosphonic acid | | Authors: | Okrasa, K, Levy, C, Leys, D, Micklefield, J. | | Deposit date: | 2009-08-17 | | Release date: | 2009-10-06 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.498 Å) | | Cite: | Structure-Guided Directed Evolution of Alkenyl and Arylmalonate Decarboxylases.
Angew.Chem.Int.Ed.Engl., 48, 2009
|
|
5HY5
 
 | |
8RA0
 
 | | Crystal structure of CysF | | Descriptor: | AMP-dependent synthetase, CITRATE ANION, TETRAETHYLENE GLYCOL | | Authors: | Levy, C.W. | | Deposit date: | 2023-11-30 | | Release date: | 2024-06-26 | | Last modified: | 2024-07-10 | | Method: | X-RAY DIFFRACTION (1.89 Å) | | Cite: | Cryptic enzymatic assembly of peptides armed with beta-lactone warheads.
Nat.Chem.Biol., 2024
|
|
6Q57
 
 | |
4Z43
 
 | | Crystal structure of Tryptophan 7-halogenase (PrnA) Mutant E450K | | Descriptor: | CHLORIDE ION, FLAVIN-ADENINE DINUCLEOTIDE, Flavin-dependent tryptophan halogenase PrnA, ... | | Authors: | Levy, C.W. | | Deposit date: | 2015-04-01 | | Release date: | 2016-01-20 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.29 Å) | | Cite: | Extending the biocatalytic scope of regiocomplementary flavin-dependent halogenase enzymes.
Chem Sci, 6, 2015
|
|
5LV9
 
 | |
5LVA
 
 | |
5FHR
 
 | |
7A9I
 
 | |
7A9J
 
 | |
5FHQ
 
 | |
3LA5
 
 | |
4LX6
 
 | |
4LX5
 
 | | X-ray crystal structure of the M6" riboswitch aptamer bound to pyrimido[4,5-d]pyrimidine-2,4-diamine (PPDA) | | Descriptor: | MAGNESIUM ION, Mutated adenine riboswitch aptamer, pyrimido[4,5-d]pyrimidine-2,4-diamine | | Authors: | Dunstan, M.S, Leys, D. | | Deposit date: | 2013-07-29 | | Release date: | 2014-07-16 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.13 Å) | | Cite: | Modular riboswitch toolsets for synthetic genetic control in diverse bacterial species.
J.Am.Chem.Soc., 136, 2014
|
|
6GKV
 
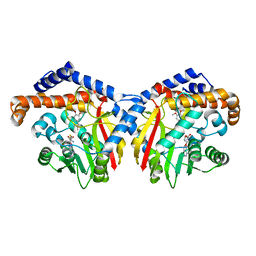 | | Crystal structure of Coclaurine N-Methyltransferase (CNMT) bound to N-methylheliamine and SAH | | Descriptor: | 6,7-dimethoxy-2-methyl-1,2,3,4-tetrahydroisoquinolin-2-ium, Coclaurine N-methyltransferase, S-ADENOSYL-L-HOMOCYSTEINE | | Authors: | Dunstan, M.S, Levy, C.W. | | Deposit date: | 2018-05-22 | | Release date: | 2018-06-06 | | Last modified: | 2024-05-15 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Structure and Biocatalytic Scope of Coclaurine N-Methyltransferase.
Angew. Chem. Int. Ed. Engl., 57, 2018
|
|
6GKY
 
 | |
6GKZ
 
 | |
