4R7N
 
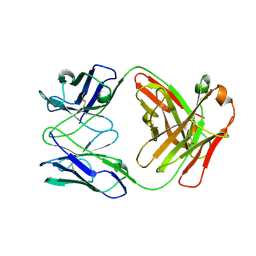 | | Fab C2E3 | | Descriptor: | Fab C2E3 Heavy chain, Fab C2E3 Light chain | | Authors: | Loyau, J, Didelot, G, Malinge, P, Ravn, U, Magistrelli, G, Depoisier, J.F, Kosco-Vilbois, M, Fischer, N, Thore, S, Rousseau, F. | | Deposit date: | 2014-08-28 | | Release date: | 2015-07-29 | | Last modified: | 2015-08-26 | | Method: | X-RAY DIFFRACTION (3.45 Å) | | Cite: | Robust Antibody-Antigen Complexes Prediction Generated by Combining Sequence Analyses, Mutagenesis, In Vitro Evolution, X-ray Crystallography and In Silico Docking.
J.Mol.Biol., 427, 2015
|
|
4R7D
 
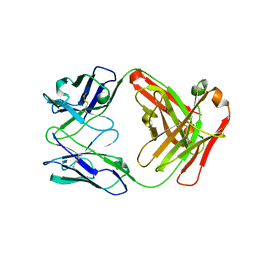 | | Fab Hu 15C1 | | Descriptor: | Fab Hu 15C1 Heavy chain, Fab Hu 15C1 Light chain | | Authors: | Loyau, J, Didelot, G, Malinge, P, Ravn, U, Magistrelli, G, Depoisier, J.F, Kosco-Vilbois, M, Fischer, N, Thore, S, Rousseau, F. | | Deposit date: | 2014-08-27 | | Release date: | 2015-07-29 | | Last modified: | 2015-08-26 | | Method: | X-RAY DIFFRACTION (2.753 Å) | | Cite: | Robust Antibody-Antigen Complexes Prediction Generated by Combining Sequence Analyses, Mutagenesis, In Vitro Evolution, X-ray Crystallography and In Silico Docking.
J.Mol.Biol., 427, 2015
|
|
3I0M
 
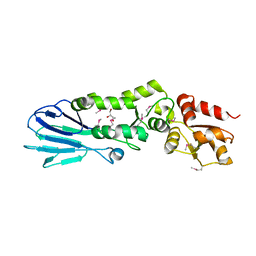 | | Structure of the S. pombe Nbs1 FHA/BRCT-repeat domain | | Descriptor: | DNA repair and telomere maintenance protein nbs1, GLYCEROL | | Authors: | Clapperton, J.A, Lloyd, J, Chapman, J.R, Jackson, S.P, Smerdon, S.J. | | Deposit date: | 2009-06-25 | | Release date: | 2009-10-13 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | A supramodular FHA/BRCT-repeat architecture mediates Nbs1 adaptor function in response to DNA damage
Cell(Cambridge,Mass.), 139, 2009
|
|
3I0N
 
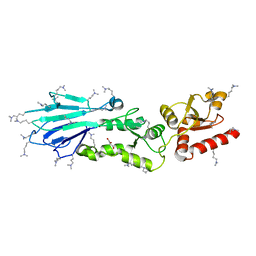 | | Structure of the S. pombe Nbs1 FHA/BRCT-repeat domain | | Descriptor: | DNA repair and telomere maintenance protein nbs1, GLYCEROL | | Authors: | Clapperton, J.A, Lloyd, J, Chapman, J.R, Jackson, S.P, Smerdon, S.J. | | Deposit date: | 2009-06-25 | | Release date: | 2009-10-13 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | A supramodular FHA/BRCT-repeat architecture mediates Nbs1 adaptor function in response to DNA damage
Cell(Cambridge,Mass.), 139, 2009
|
|
3UOT
 
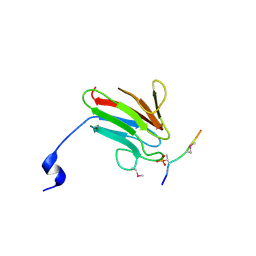 | | Crystal Structure of MDC1 FHA Domain in Complex with a Phosphorylated Peptide from the MDC1 N-terminus | | Descriptor: | Mediator of DNA damage checkpoint protein 1 | | Authors: | Clapperton, J.A, Lloyd, J, Haire, L.F, Li, J, Smerdon, S.J. | | Deposit date: | 2011-11-17 | | Release date: | 2011-12-28 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | The molecular basis of ATM-dependent dimerization of the Mdc1 DNA damage checkpoint mediator.
Nucleic Acids Res., 40, 2012
|
|
3UN0
 
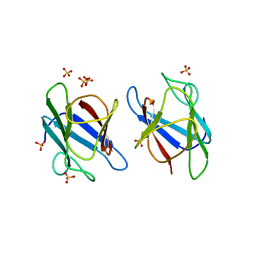 | | Crystal Structure of MDC1 FHA Domain | | Descriptor: | Mediator of DNA damage checkpoint protein 1, SULFATE ION | | Authors: | Clapperton, J.A, Lloyd, J, Haire, L.F, Li, J, Smerdon, S.J. | | Deposit date: | 2011-11-15 | | Release date: | 2011-12-28 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | The molecular basis of ATM-dependent dimerization of the Mdc1 DNA damage checkpoint mediator.
Nucleic Acids Res., 40, 2012
|
|
1DDJ
 
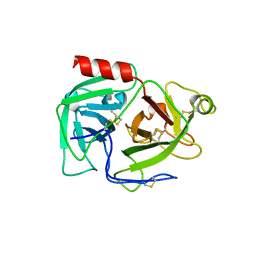 | | CRYSTAL STRUCTURE OF HUMAN PLASMINOGEN CATALYTIC DOMAIN | | Descriptor: | PLASMINOGEN | | Authors: | Wang, X, Terzyan, S, Tang, J, Loy, J, Lin, X, Zhang, X. | | Deposit date: | 1999-11-10 | | Release date: | 2000-02-18 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Human plasminogen catalytic domain undergoes an unusual conformational change upon activation.
J.Mol.Biol., 295, 2000
|
|
7D5L
 
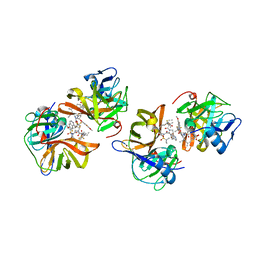 | | Discovery of BMS-986144, a Third Generation, Pan Genotype NS3/4A Protease Inhibitor for the Treatment of Hepatitis C Virus Infection | | Descriptor: | NS3/4A Protease, ZINC ION, [1,1,1-tris(fluoranyl)-2-methyl-propan-2-yl] ~{N}-[(1~{S},4~{R},6~{S},7~{Z},11~{R},13~{R},14~{S},18~{R})-13-ethyl-18-(7-fluoranyl-6-methoxy-isoquinolin-1-yl)oxy-11-methyl-4-[(1-methylcyclopropyl)sulfonylcarbamoyl]-2,15-bis(oxidanylidene)-3,16-diazatricyclo[14.3.0.0^{4,6}]nonadec-7-en-14-yl]carbamate | | Authors: | Ghosh, K, Anumula, R, Kumar, A. | | Deposit date: | 2020-09-26 | | Release date: | 2020-12-16 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Discovery of BMS-986144, a Third-Generation, Pan-Genotype NS3/4A Protease Inhibitor for the Treatment of Hepatitis C Virus Infection.
J.Med.Chem., 63, 2020
|
|
8VDJ
 
 | | Crystal structure of SARS-CoV-2 3CL protease (3CLpro) as a covalent complex with EDP-235 | | Descriptor: | 3C-like proteinase nsp5, 4,6,7-trifluoro-N-{(2S)-1-[(3R,5'R)-5'-(iminomethyl)-2-oxo-1,2-dihydrospiro[indole-3,3'-pyrrolidin]-1'-yl]-4-methyl-1-oxopentan-2-yl}-N-methyl-1H-indole-2-carboxamide, THIOCYANATE ION | | Authors: | Cade, I.A, Rhodin, M.H.J. | | Deposit date: | 2023-12-15 | | Release date: | 2024-08-14 | | Method: | X-RAY DIFFRACTION (2.002 Å) | | Cite: | The small molecule inhibitor of SARS-CoV-2 3CLpro EDP-235 prevents viral replication and transmission in vivo.
Nat Commun, 15, 2024
|
|
6N7B
 
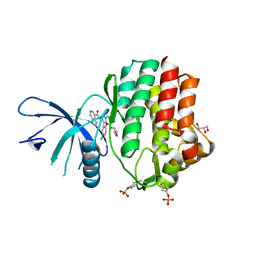 | | Structure of the human JAK1 kinase domain with compound 38 | | Descriptor: | GLYCEROL, N-[3-(5-chloro-2-methoxyphenyl)-1-methyl-1H-pyrazol-4-yl]-1H-pyrazolo[4,3-c]pyridine-7-carboxamide, Tyrosine-protein kinase JAK1 | | Authors: | Lupardus, P.J, Brown, D. | | Deposit date: | 2018-11-27 | | Release date: | 2019-04-24 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.81 Å) | | Cite: | Discovery of a class of highly potent Janus Kinase 1/2 (JAK1/2) inhibitors demonstrating effective cell-based blockade of IL-13 signaling.
Bioorg.Med.Chem.Lett., 29, 2019
|
|
6N79
 
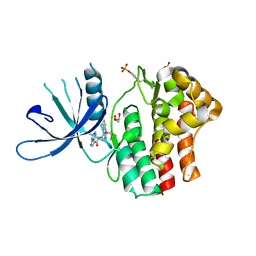 | | Structure of the human JAK1 kinase domain with compound 20 | | Descriptor: | GLYCEROL, N-{5-[5-chloro-2-(difluoromethoxy)phenyl]-1H-pyrazol-4-yl}pyrazolo[1,5-a]pyrimidine-3-carboxamide, Tyrosine-protein kinase JAK1 | | Authors: | Lupardus, P.J, Brown, D. | | Deposit date: | 2018-11-27 | | Release date: | 2019-04-24 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.27 Å) | | Cite: | Discovery of a class of highly potent Janus Kinase 1/2 (JAK1/2) inhibitors demonstrating effective cell-based blockade of IL-13 signaling.
Bioorg.Med.Chem.Lett., 29, 2019
|
|
6N7C
 
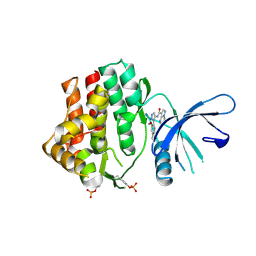 | | Structure of the human JAK1 kinase domain with compound 56 | | Descriptor: | GLYCEROL, N-[5-(3-methoxynaphthalen-2-yl)-1H-pyrazol-4-yl]pyrazolo[1,5-a]pyrimidine-3-carboxamide, Tyrosine-protein kinase JAK1 | | Authors: | Lupardus, P.J, Brown, D. | | Deposit date: | 2018-11-27 | | Release date: | 2019-04-24 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.69 Å) | | Cite: | Discovery of a class of highly potent Janus Kinase 1/2 (JAK1/2) inhibitors demonstrating effective cell-based blockade of IL-13 signaling.
Bioorg.Med.Chem.Lett., 29, 2019
|
|
6N77
 
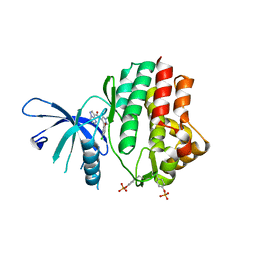 | | Structure of the human JAK1 kinase domain with compound 15 | | Descriptor: | GLYCEROL, N-[3-(5-chloro-2-methoxyphenyl)-1-methyl-1H-pyrazol-4-yl]pyrazolo[1,5-a]pyrimidine-3-carboxamide, Tyrosine-protein kinase JAK1 | | Authors: | Lupardus, P.J, Brown, D. | | Deposit date: | 2018-11-27 | | Release date: | 2019-04-24 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.64 Å) | | Cite: | Discovery of a class of highly potent Janus Kinase 1/2 (JAK1/2) inhibitors demonstrating effective cell-based blockade of IL-13 signaling.
Bioorg.Med.Chem.Lett., 29, 2019
|
|
6N7D
 
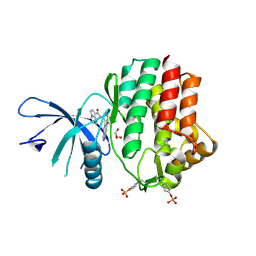 | | Structure of the human JAK1 kinase domain with compound 54 | | Descriptor: | GLYCEROL, N-[5-(6-methoxy-1H-indazol-5-yl)-1H-pyrazol-4-yl]pyrazolo[1,5-a]pyrimidine-3-carboxamide, Tyrosine-protein kinase JAK1 | | Authors: | Lupardus, P.J, Brown, D. | | Deposit date: | 2018-11-27 | | Release date: | 2019-04-24 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.78 Å) | | Cite: | Discovery of a class of highly potent Janus Kinase 1/2 (JAK1/2) inhibitors demonstrating effective cell-based blockade of IL-13 signaling.
Bioorg.Med.Chem.Lett., 29, 2019
|
|
6N78
 
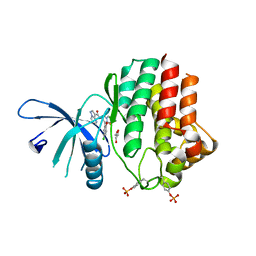 | | Structure of the human JAK1 kinase domain with compound 21 | | Descriptor: | GLYCEROL, N-{3-[5-chloro-2-(difluoromethoxy)phenyl]-1-methyl-1H-pyrazol-4-yl}pyrazolo[1,5-a]pyrimidine-3-carboxamide, Tyrosine-protein kinase JAK1 | | Authors: | Lupardus, P.J, Brown, D. | | Deposit date: | 2018-11-27 | | Release date: | 2019-04-24 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.83 Å) | | Cite: | Discovery of a class of highly potent Janus Kinase 1/2 (JAK1/2) inhibitors demonstrating effective cell-based blockade of IL-13 signaling.
Bioorg.Med.Chem.Lett., 29, 2019
|
|
6N7A
 
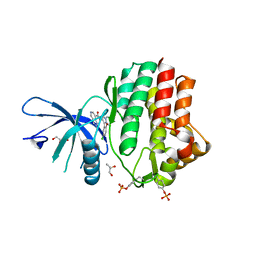 | | Structure of the human JAK1 kinase domain with compound 39 | | Descriptor: | GLYCEROL, N-[3-(5-chloro-2-methoxyphenyl)-1-methyl-1H-pyrazol-4-yl]-2-methyl-2H-pyrazolo[4,3-c]pyridine-7-carboxamide, Tyrosine-protein kinase JAK1 | | Authors: | Lupardus, P.J, Brown, D. | | Deposit date: | 2018-11-27 | | Release date: | 2019-04-24 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.33 Å) | | Cite: | Discovery of a class of highly potent Janus Kinase 1/2 (JAK1/2) inhibitors demonstrating effective cell-based blockade of IL-13 signaling.
Bioorg.Med.Chem.Lett., 29, 2019
|
|
3MAC
 
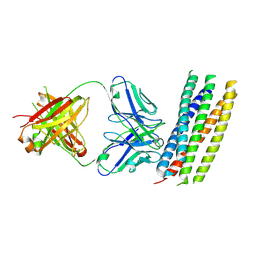 | | crystal structure of GP41-derived protein complexed with fab 8062 | | Descriptor: | Fab8062, Transmembrane glycoprotein | | Authors: | Li, M, Gustchina, E, Louis, J, Gustchina, A, Wlodawer, A, Clore, M. | | Deposit date: | 2010-03-23 | | Release date: | 2010-12-08 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structural Basis of HIV-1 Neutralization by Affinity Matured Fabs Directed against the Internal Trimeric Coiled-Coil of gp41.
Plos Pathog., 6, 2010
|
|
3MA9
 
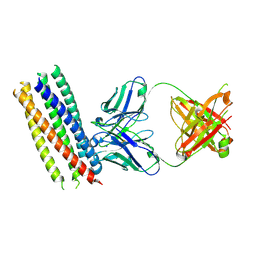 | | Crystal structure of gp41 derived protein complexed with fab 8066 | | Descriptor: | Fab8066 FAB ANTIBODY FRAGMENT, Heavy Chain, Light Chain, ... | | Authors: | Li, M, Gustchina, E, Louis, J, Gustchina, A, Wlodawer, A, Clore, M. | | Deposit date: | 2010-03-23 | | Release date: | 2010-12-01 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Structural Basis of HIV-1 Neutralization by Affinity Matured Fabs Directed against the Internal Trimeric Coiled-Coil of gp41.
Plos Pathog., 6, 2010
|
|
9F7K
 
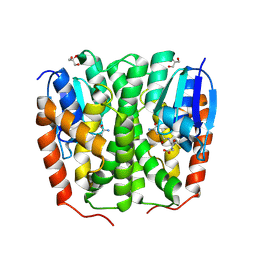 | | Glutathione transferase epsilon 1 from Drosophila melanogaster in complex with glutathione | | Descriptor: | GH14654p, GLUTATHIONE, GLYCEROL, ... | | Authors: | Didierjean, C, Schwartz, M, Neiers, F. | | Deposit date: | 2024-05-03 | | Release date: | 2024-09-04 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structural and Thermodynamic Insights into Dimerization Interfaces of Drosophila Glutathione Transferases.
Biomolecules, 14, 2024
|
|
6RNX
 
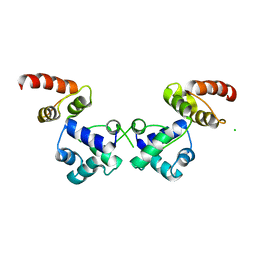 | | Crystal structure of the essential repressor DdrO from radiation-resistant Deinococcus bacteria (Deinococcus deserti) | | Descriptor: | CHLORIDE ION, HTH-type transcriptional regulator DdrOC | | Authors: | Arnoux, P, Siponen, M.I, Pignol, D, De Groot, A, Blanchard, L. | | Deposit date: | 2019-05-09 | | Release date: | 2019-10-09 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.84 Å) | | Cite: | Crystal structure of the transcriptional repressor DdrO: insight into the metalloprotease/repressor-controlled radiation response in Deinococcus.
Nucleic Acids Res., 47, 2019
|
|
6RNZ
 
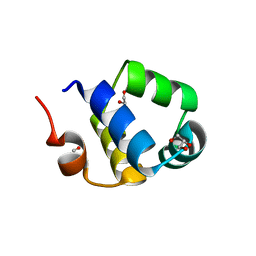 | | Crystal structure of the N-terminal HTH DNA-binding domain of the essential repressor DdrO from radiation-resistant Deinococcus bacteria (Deinococcus deserti) | | Descriptor: | GLYCEROL, HTH-type transcriptional regulator DdrOC | | Authors: | Arnoux, P, Siponen, M.I, Pignol, D, Brandelet, G, De Groot, A, Blanchard, L. | | Deposit date: | 2019-05-10 | | Release date: | 2019-10-09 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.35 Å) | | Cite: | Crystal structure of the transcriptional repressor DdrO: insight into the metalloprotease/repressor-controlled radiation response in Deinococcus.
Nucleic Acids Res., 47, 2019
|
|
6RO6
 
 | | Crystal structure of the C-terminal dimerization domain of the essential repressor DdrO from radiation-resistant Deinococcus bacteria (Deinococcus deserti) | | Descriptor: | HTH-type transcriptional regulator DdrOC, SULFATE ION | | Authors: | Pignol, D, Arnoux, P, Siponen, M.I, Brandelet, G, De Groot, A, Blanchard, L. | | Deposit date: | 2019-05-10 | | Release date: | 2019-10-09 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.41 Å) | | Cite: | Crystal structure of the transcriptional repressor DdrO: insight into the metalloprotease/repressor-controlled radiation response in Deinococcus.
Nucleic Acids Res., 47, 2019
|
|
6RMQ
 
 | | Crystal structure of a selenomethionine-substituted A70M I84M mutant of the essential repressor DdrO from radiation resistant-Deinococcus bacteria (Deinococcus deserti) | | Descriptor: | HTH-type transcriptional regulator DdrOC | | Authors: | Arnoux, P, Siponen, M.I, Pignol, D, De Groot, A, Blanchard, L. | | Deposit date: | 2019-05-07 | | Release date: | 2019-10-09 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Crystal structure of the transcriptional repressor DdrO: insight into the metalloprotease/repressor-controlled radiation response in Deinococcus.
Nucleic Acids Res., 47, 2019
|
|
