6F22
 
 | | Complex between MTH1 and compound 29 (a 4-amino-2,7-diazaindole derivative) | | Descriptor: | (3~{S})-3-phenyl-4-(2~{H}-pyrazolo[3,4-b]pyridin-4-yl)morpholine, 7,8-dihydro-8-oxoguanine triphosphatase, SULFATE ION | | Authors: | Viklund, J, Talagas, A, Tresaugues, L, Andersson, M, Ericsson, U, Forsblom, R, Ginman, T, Hallberg, K, Lindstrom, J, Persson, L, Silvander, C, Rahm, F. | | Deposit date: | 2017-11-23 | | Release date: | 2018-03-07 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Creation of a Novel Class of Potent and Selective MutT Homologue 1 (MTH1) Inhibitors Using Fragment-Based Screening and Structure-Based Drug Design.
J. Med. Chem., 61, 2018
|
|
6F1X
 
 | | Complex between MTH1 and compound 7 (a 7-azaindole-2-amide derivative) | | Descriptor: | 4-(3-chlorophenyl)-~{N}-ethyl-1~{H}-pyrrolo[2,3-b]pyridine-2-carboxamide, 7,8-dihydro-8-oxoguanine triphosphatase, SULFATE ION | | Authors: | Viklund, J, Talagas, A, Tresaugues, L, Andersson, M, Ericsson, U, Forsblom, R, Ginman, T, Hallberg, K, Lindstrom, J, Persson, L, Silvander, C, Rahm, F. | | Deposit date: | 2017-11-23 | | Release date: | 2018-03-07 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Creation of a Novel Class of Potent and Selective MutT Homologue 1 (MTH1) Inhibitors Using Fragment-Based Screening and Structure-Based Drug Design.
J. Med. Chem., 61, 2018
|
|
5NHY
 
 | | BAY-707 in complex with MTH1 | | Descriptor: | 7,8-dihydro-8-oxoguanine triphosphatase, GLYCEROL, SULFATE ION, ... | | Authors: | Ellermann, M, Eheim, A, Giese, A, Bunse, S, Nowak-Reppel, K, Neuhaus, R, Weiske, J, Quanz, M, Glasauer, A, Meyer, H, Queisser, N, Irlbacher, H, Bader, B, Rahm, F, Viklund, J, Andersson, M, Ericsson, U, Ginman, T, Forsblom, R, Lindstrom, J, Silvander, C, Tresaugues, L, Gorjanacz, M. | | Deposit date: | 2017-03-22 | | Release date: | 2017-07-19 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.72 Å) | | Cite: | Novel Class of Potent and Cellularly Active Inhibitors Devalidates MTH1 as Broad-Spectrum Cancer Target.
ACS Chem. Biol., 12, 2017
|
|
6F23
 
 | | Complex between MTH1 and compound 16 (a 4-amino-7-azaindole derivative) | | Descriptor: | 4-[(2~{R})-2-phenylpyrrolidin-1-yl]-1~{H}-pyrrolo[2,3-b]pyridine, 7,8-dihydro-8-oxoguanine triphosphatase, GLYCEROL, ... | | Authors: | Viklund, J, Tresaugues, L, Talagas, A, Andersson, M, Ericsson, U, Forsblom, R, Ginman, T, Hallberg, K, Lindstrom, J, Persson, L, Silvander, C, Rahm, F. | | Deposit date: | 2017-11-23 | | Release date: | 2018-03-07 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.84 Å) | | Cite: | Creation of a Novel Class of Potent and Selective MutT Homologue 1 (MTH1) Inhibitors Using Fragment-Based Screening and Structure-Based Drug Design.
J. Med. Chem., 61, 2018
|
|
6F20
 
 | | Complex between MTH1 and compound 1 (a 7-azaindole-4-ester derivative) | | Descriptor: | 7,8-dihydro-8-oxoguanine triphosphatase, ACETATE ION, Ethyl 1H-pyrrolo[2,3-b]pyridine-4-carboxylate, ... | | Authors: | Viklund, J, Talagas, A, Tresaugues, L, Andersson, M, Ericsson, U, Forsblom, R, Ginman, T, Hallberg, K, Lindstrom, J, Persson, L, Silvander, C, Rahm, F. | | Deposit date: | 2017-11-23 | | Release date: | 2018-03-07 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Creation of a Novel Class of Potent and Selective MutT Homologue 1 (MTH1) Inhibitors Using Fragment-Based Screening and Structure-Based Drug Design.
J. Med. Chem., 61, 2018
|
|
8RXR
 
 | | Crystal structure of VPS34 in complex with inhibitor SB02024 | | Descriptor: | 4-[(3R)-3-methylmorpholin-4-yl]-2-[(2R)-2-(trifluoromethyl)piperidin-1-yl]-3H-pyridin-6-one, DI(HYDROXYETHYL)ETHER, DIMETHYL SULFOXIDE, ... | | Authors: | Tresaugues, L, Yu, Y, Bogdan, M, Parpal, S, Silvander, C, Lindstrom, J, Simeon, J, Timson, M.J, Al-Hashimi, H, Smith, B.D, Flynn, D.L, Viklund, J, Martinsson, J, De Milito, A, Andersson, M. | | Deposit date: | 2024-02-07 | | Release date: | 2024-03-20 | | Last modified: | 2024-08-21 | | Method: | X-RAY DIFFRACTION (2.06 Å) | | Cite: | Combining VPS34 inhibitors with STING agonists enhances type I interferon signaling and anti-tumor efficacy.
Mol Oncol, 18, 2024
|
|
5UG3
 
 | | NMR SOLUTION STRUCTURE OF ALPHA-CONOTOXIN GID MUTANT A10V | | Descriptor: | Alpha-conotoxin GID | | Authors: | Hussein, A.K, Leffler, A.E, Zebroski, H.A, Powell, S.R, Kuryatov, A, Filipenko, P, Gorson, J, Heizmann, A, Lyskov, S, Nicke, A, Lindstrom, J, Rudy, B, Bonneau, R, Holford, M, Poget, S.F. | | Deposit date: | 2017-01-06 | | Release date: | 2017-09-06 | | Last modified: | 2023-06-14 | | Method: | SOLUTION NMR | | Cite: | Discovery of peptide ligands through docking and virtual screening at nicotinic acetylcholine receptor homology models.
Proc. Natl. Acad. Sci. U.S.A., 114, 2017
|
|
5UG5
 
 | | NMR SOLUTION STRUCTURE OF THE ALPHA-CONOTOXIN GID MUTANT V13Y | | Descriptor: | Alpha-conotoxin GID | | Authors: | Hussein, A, Leffler, A.E, Kuryatov, A, Zebroski, H.A, Powell, S.R, Filipenko, P, Gorson, J, Heizmann, A, Lyskov, S, Nicke, A, Lindstrom, J, Rudy, B, Bonneau, R, Holford, M, Poget, S.F. | | Deposit date: | 2017-01-06 | | Release date: | 2017-09-06 | | Last modified: | 2023-06-14 | | Method: | SOLUTION NMR | | Cite: | Discovery of peptide ligands through docking and virtual screening at nicotinic acetylcholine receptor homology models.
Proc. Natl. Acad. Sci. U.S.A., 114, 2017
|
|
4B1D
 
 | | New Aminoimidazoles as BACE-1 Inhibitors: From Rational Design to Ab- lowering in Brain | | Descriptor: | (2S)-2-(4-methoxy-3,5-dimethylphenyl)-5-methyl-2-(3-pyrimidin-5-ylphenyl)-2H-imidazol-4-amine, ACETATE ION, BETA-SECRETASE 1 | | Authors: | Rahm, F, Blid, J, Ginman, T, Karlstrom, S, Kihlstrom, J, Kolmodin, K, Lindstrom, J, von Berg, S, von Kieseritzky, F, Slivo, C, Swahn, B, Viklund, J, Olsson, L, Johansson, P, Eketjall, S, Falting, J, Jeppsson, F, Stromberg, K, Janson, J, Gravenfors, Y. | | Deposit date: | 2012-07-10 | | Release date: | 2012-10-10 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | New aminoimidazoles as beta-secretase (BACE-1) inhibitors showing amyloid-beta (A beta ) lowering in brain.
J. Med. Chem., 55, 2012
|
|
4B1E
 
 | | New Aminoimidazoles as BACE-1 Inhibitors: From Rational Design to Ab- lowering in Brain | | Descriptor: | (2R)-2-methyl-5-phenyl-2-(3-pyridin-3-ylphenyl)-2,3-dihydro-1H-imidazol-4-amine, BETA-SECRETASE 1 | | Authors: | Rahm, F, Blid, J, Ginman, T, Karlstrom, S, Kihlstrom, J, Kolmodin, K, Lindstrom, J, von Berg, S, von Kieseritzky, F, Slivo, C, Swahn, B, Viklund, J, Olsson, L, Johansson, P, Eketjall, S, Falting, J, Jeppsson, F, Stromberg, K, Janson, J, Gravenfors, Y. | | Deposit date: | 2012-07-10 | | Release date: | 2012-10-10 | | Last modified: | 2018-02-14 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | New aminoimidazoles as beta-secretase (BACE-1) inhibitors showing amyloid-beta (A beta ) lowering in brain.
J. Med. Chem., 55, 2012
|
|
4B1C
 
 | | New Aminoimidazoles as BACE-1 Inhibitors: From Rational Design to Ab- lowering in Brain | | Descriptor: | (2R)-2-cyclopropyl-5-methyl-2-[3-(5-prop-1-yn-1-ylpyridin-3-yl)phenyl]-2H-imidazol-4-amine, BETA-SECRETASE 1, DIMETHYL SULFOXIDE | | Authors: | Rahm, F, Blid, J, Ginman, T, Karlstrom, S, Kihlstrom, J, Kolmodin, K, Lindstrom, J, von Berg, S, von Kieseritzky, F, Slivo, C, Swahn, B, Viklund, J, Olsson, L, Johansson, P, Eketjall, S, Falting, J, Jeppsson, F, Stromberg, K, Janson, J, Gravenfors, Y. | | Deposit date: | 2012-07-10 | | Release date: | 2012-10-10 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | New aminoimidazoles as beta-secretase (BACE-1) inhibitors showing amyloid-beta (A beta ) lowering in brain.
J. Med. Chem., 55, 2012
|
|
4AZY
 
 | | Design and Synthesis of BACE1 Inhibitors with In Vivo Brain Reduction of beta-Amyloid Peptides (COMPOUND 10) | | Descriptor: | (1S)-4-fluoro-1-(4-fluoro-3-pyrimidin-5-ylphenyl)-1-[2-(trifluoromethyl)pyridin-4-yl]-1H-isoindol-3-amine, ACETATE ION, BETA-SECRETASE 1, ... | | Authors: | Swahn, B.M, Kolmodin, K, Karlstrom, S, von Berg, S, Soderman, P, Holenz, J, Berg, S, Lindstrom, J, Sundstrom, M, Turek, D, Kihlstrom, J, Slivo, C, Andersson, L, Pyring, D, Ohberg, L, Kers, A, Bogar, K, Bergh, M, Olsson, L.L, Janson, J, Eketjall, S, Georgievska, B, Jeppsson, F, Falting, J. | | Deposit date: | 2012-06-27 | | Release date: | 2012-10-17 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.79 Å) | | Cite: | Design and synthesis of beta-site amyloid precursor protein cleaving enzyme (BACE1) inhibitors with in vivo brain reduction of beta-amyloid peptides.
J. Med. Chem., 55, 2012
|
|
4ACU
 
 | | Aminoimidazoles as BACE-1 Inhibitors. X-RAY CRYSTAL STRUCTURE OF BETA SECRETASE COMPLEXED WITH COMPOUND 14 | | Descriptor: | (8S)-3,3-DIFLUORO-8-(2'-FLUORO-3'-METHOXYBIPHENYL-3-YL)-8-PYRIDIN-4-YL-2,3,4,8-TETRAHYDROIMIDAZO[1,5-A]PYRIMIDIN-6-AMINE, ACETATE ION, BETA-SECRETASE 1 | | Authors: | Swahn, B, Holenz, J, Kihlstrom, J, Kolmodin, K, Lindstrom, J, Plobeck, N, Rotticci, D, Sehgelmeble, F, Sundstrom, M, von Berg, S, Falting, J, Georgievska, B, Gustavsson, S, Neelissen, J, Ek, M, Olsson, L.L, Berg, S. | | Deposit date: | 2011-12-19 | | Release date: | 2012-02-29 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Aminoimidazoles as BACE-1 inhibitors: the challenge to achieve in vivo brain efficacy.
Bioorg. Med. Chem. Lett., 22, 2012
|
|
4ACX
 
 | | Aminoimidazoles as BACE-1 Inhibitors. X-RAY CRYSTAL STRUCTURE OF BETA SECRETASE COMPLEXED WITH COMPOUND 23 | | Descriptor: | (8R)-8-[4-(DIFLUOROMETHOXY)PHENYL]-3,3-DIFLUORO-8-[3-(3-METHOXYPROP-1-YN-1-YL)PHENYL]-2,3,4,8-TETRAHYDROIMIDAZO[1,5-A]PYRIMIDIN-6-AMINE, ACETATE ION, BETA-SECRETASE 1 | | Authors: | Swahn, B, Holenz, J, Kihlstrom, J, Kolmodin, K, Lindstrom, J, Plobeck, N, Rotticci, D, Sehgelmeble, F, Sundstrom, M, von Berg, S, Falting, J, Georgievska, B, Gustavsson, S, Neelissen, J, Ek, M, Olsson, L.L, Berg, S. | | Deposit date: | 2011-12-20 | | Release date: | 2012-02-29 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Aminoimidazoles as BACE-1 inhibitors: the challenge to achieve in vivo brain efficacy.
Bioorg. Med. Chem. Lett., 22, 2012
|
|
4B00
 
 | | Design and Synthesis of BACE1 Inhibitors with In Vivo Brain Reduction of beta-Amyloid Peptides (COMPOUND (R)-41) | | Descriptor: | 5-{(1R)-3-amino-4-fluoro-1-[3-(5-prop-1-yn-1-ylpyridin-3-yl)phenyl]-1H-isoindol-1-yl}-1-ethyl-3-methylpyridin-2(1H)-one, ACETATE ION, BETA-SECRETASE 1 | | Authors: | Swahn, B.M, Kolmodin, K, Karlstrom, S, von Berg, S, Soderman, P, Holenz, J, Berg, S, Lindstrom, J, Sundstrom, M, Turek, D, Kihlstrom, J, Slivo, C, Andersson, L, Pyring, D, Ohberg, L, Kers, A, Bogar, K, Bergh, M, Olsson, L.L, Janson, J, Eketjall, S, Georgievska, B, Jeppsson, F, Falting, J. | | Deposit date: | 2012-06-27 | | Release date: | 2012-10-17 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.83 Å) | | Cite: | Design and synthesis of beta-site amyloid precursor protein cleaving enzyme (BACE1) inhibitors with in vivo brain reduction of beta-amyloid peptides.
J. Med. Chem., 55, 2012
|
|
1KW7
 
 | | METHIONINE CORE MUTANT OF T4 LYSOZYME | | Descriptor: | 2-HYDROXYETHYL DISULFIDE, CHLORIDE ION, LYSOZYME | | Authors: | Gassner, N.C, Baase, W.A, Mooers, B.H, Busam, R.D, Weaver, L.H, Lindstrom, J.D, Quillin, M.L, Matthews, B.W. | | Deposit date: | 2002-01-28 | | Release date: | 2003-06-03 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.89 Å) | | Cite: | Multiple methionine substitutions are tolerated in T4 lysozyme and have coupled effects on folding and stability
BIOPHYS.CHEM., 100, 2003
|
|
1KW5
 
 | | METHIONINE CORE MUTANT OF T4 LYSOZYME | | Descriptor: | 2-HYDROXYETHYL DISULFIDE, CHLORIDE ION, LYSOZYME | | Authors: | Gassner, N.C, Baase, W.A, Mooers, B.H, Busam, R.D, Weaver, L.H, Lindstrom, J.D, Quillin, M.L, Matthews, B.W. | | Deposit date: | 2002-01-28 | | Release date: | 2003-06-03 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Multiple methionine substitutions are tolerated in T4 lysozyme and have coupled effects on folding and stability
BIOPHYS.CHEM., 100, 2003
|
|
1CVK
 
 | | T4 LYSOZYME MUTANT L118A | | Descriptor: | 2-HYDROXYETHYL DISULFIDE, CHLORIDE ION, LYSOZYME | | Authors: | Gassner, N.C, Baase, W.A, Lindstrom, J, Lu, J, Matthews, B.W. | | Deposit date: | 1999-08-23 | | Release date: | 1999-11-10 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Methionine and alanine substitutions show that the formation of wild-type-like structure in the carboxy-terminal domain of T4 lysozyme is a rate-limiting step in folding.
Biochemistry, 38, 1999
|
|
1CV4
 
 | | T4 LYSOZYME MUTANT L118M | | Descriptor: | 2-HYDROXYETHYL DISULFIDE, CHLORIDE ION, LYSOZYME | | Authors: | Gassner, N.C, Baase, W.A, Lindstrom, J, Lu, J, Matthews, B.W. | | Deposit date: | 1999-08-22 | | Release date: | 1999-11-10 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Methionine and alanine substitutions show that the formation of wild-type-like structure in the carboxy-terminal domain of T4 lysozyme is a rate-limiting step in folding.
Biochemistry, 38, 1999
|
|
1CV6
 
 | | T4 LYSOZYME MUTANT V149M | | Descriptor: | 2-HYDROXYETHYL DISULFIDE, CHLORIDE ION, LYSOZYME | | Authors: | Gassner, N.C, Baase, W.A, Lindstrom, J, Lu, J, Matthews, B.W. | | Deposit date: | 1999-08-22 | | Release date: | 1999-11-10 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Methionine and alanine substitutions show that the formation of wild-type-like structure in the carboxy-terminal domain of T4 lysozyme is a rate-limiting step in folding.
Biochemistry, 38, 1999
|
|
1KY0
 
 | | METHIONINE CORE MUTANT OF T4 LYSOZYME | | Descriptor: | 2-HYDROXYETHYL DISULFIDE, CHLORIDE ION, LYSOZYME | | Authors: | Gassner, N.C, Baase, W.A, Mooers, B.H, Busam, R.D, Weaver, L.H, Lindstrom, J.D, Quillin, M.L, Matthews, B.W. | | Deposit date: | 2002-02-01 | | Release date: | 2003-06-03 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.97 Å) | | Cite: | Multiple methionine substitutions are tolerated in T4 lysozyme and have coupled effects on folding and stability
BIOPHYS.CHEM., 100, 2003
|
|
1L0J
 
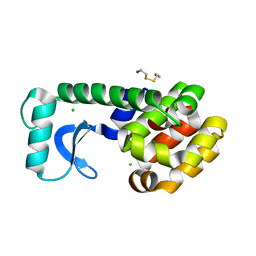 | | METHIONINE CORE MUTANT OF T4 LYSOZYME | | Descriptor: | 2-HYDROXYETHYL DISULFIDE, CHLORIDE ION, LYSOZYME | | Authors: | Gassner, N.C, Baase, W.A, Mooers, B.H, Busam, R.D, Weaver, L.H, Lindstrom, J.D, Quillin, M.L, Matthews, B.W. | | Deposit date: | 2002-02-11 | | Release date: | 2003-06-03 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.98 Å) | | Cite: | Multiple methionine substitutions are tolerated in T4 lysozyme and have coupled effects on folding and stability
BIOPHYS.CHEM., 100, 2003
|
|
1KY1
 
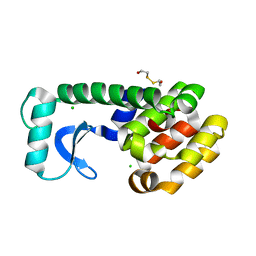 | | METHIONINE CORE MUTANT OF T4 LYSOZYME | | Descriptor: | 2-HYDROXYETHYL DISULFIDE, CHLORIDE ION, LYSOZYME | | Authors: | Gassner, N.C, Baase, W.A, Mooers, B.H, Busam, R.D, Weaver, L.H, Lindstrom, J.D, Quillin, M.L, Matthews, B.W. | | Deposit date: | 2002-02-01 | | Release date: | 2003-06-03 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Multiple methionine substitutions are tolerated in T4 lysozyme and have coupled effects on folding and stability
BIOPHYS.CHEM., 100, 2003
|
|
1L0K
 
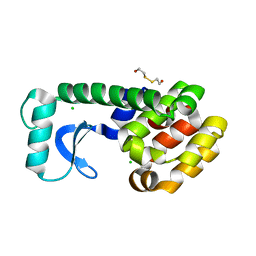 | | METHIONINE CORE MUTANT OF T4 LYSOZYME | | Descriptor: | 2-HYDROXYETHYL DISULFIDE, CHLORIDE ION, LYSOZYME | | Authors: | Gassner, N.C, Baase, W.A, Mooers, B.H, Busam, R.D, Weaver, L.H, Lindstrom, J.D, Quillin, M.L, Matthews, B.W. | | Deposit date: | 2002-02-11 | | Release date: | 2003-06-03 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.02 Å) | | Cite: | Multiple methionine substitutions are tolerated in T4 lysozyme and have coupled effects on folding and stability
BIOPHYS.CHEM., 100, 2003
|
|
1QSQ
 
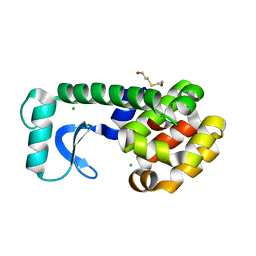 | | CAVITY CREATING MUTATION | | Descriptor: | 2-HYDROXYETHYL DISULFIDE, CHLORIDE ION, LYSOZYME | | Authors: | Gassner, N.C, Baase, W.A, Lindstrom, J, Matthews, B.W. | | Deposit date: | 1999-06-22 | | Release date: | 1999-06-29 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Methionine and alanine substitutions show that the formation of wild-type-like structure in the carboxy-terminal domain of T4 lysozyme is a rate-limiting step in folding.
Biochemistry, 38, 1999
|
|
