7D84
 
 | | 34-fold symmetry Salmonella S ring formed by full-length FliF | | Descriptor: | Flagellar M-ring protein | | Authors: | Kawamoto, A, Miyata, T, Makino, F, Kinoshita, M, Minamino, T, Imada, K, Kato, T, Namba, K. | | Deposit date: | 2020-10-07 | | Release date: | 2021-05-19 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (3.7 Å) | | Cite: | Native flagellar MS ring is formed by 34 subunits with 23-fold and 11-fold subsymmetries.
Nat Commun, 12, 2021
|
|
7YVQ
 
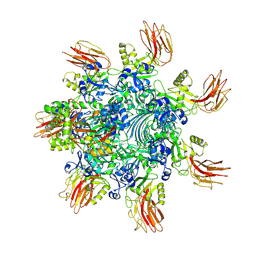 | | Complex structure of Clostridioides difficile binary toxin folded CDTa-bound CDTb-pore (short). | | Descriptor: | ADP-ribosylating binary toxin binding subunit CdtB, ADP-ribosylating binary toxin enzymatic subunit CdtA, CALCIUM ION | | Authors: | Yamada, T, Kawamoto, A, Yoshida, T, Sato, Y, Kato, T, Tsuge, H. | | Deposit date: | 2022-08-19 | | Release date: | 2022-10-26 | | Last modified: | 2024-07-03 | | Method: | ELECTRON MICROSCOPY (3.18 Å) | | Cite: | Cryo-EM structures of the translocational binary toxin complex CDTa-bound CDTb-pore from Clostridioides difficile.
Nat Commun, 13, 2022
|
|
7YVS
 
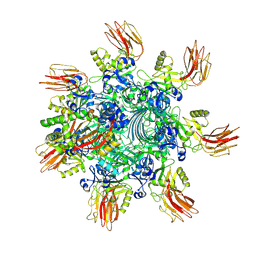 | | Complex structure of Clostridioides difficile binary toxin unfolded CDTa-bound CDTb-pore (short). | | Descriptor: | ADP-ribosylating binary toxin binding subunit CdtB, ADP-ribosylating binary toxin enzymatic subunit CdtA, CALCIUM ION | | Authors: | Yamada, T, Kawamoto, A, Yoshida, T, Sato, Y, Kato, T, Tsuge, H. | | Deposit date: | 2022-08-19 | | Release date: | 2022-10-26 | | Last modified: | 2024-07-03 | | Method: | ELECTRON MICROSCOPY (2.8 Å) | | Cite: | Cryo-EM structures of the translocational binary toxin complex CDTa-bound CDTb-pore from Clostridioides difficile.
Nat Commun, 13, 2022
|
|
7BWM
 
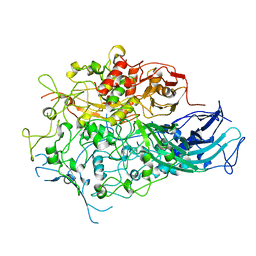 | |
7VNN
 
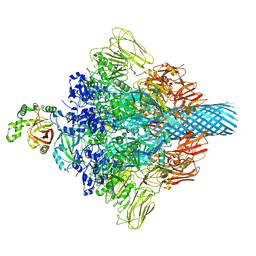 | | Complex structure of Clostridioides difficile enzymatic component (CDTa) and binding component (CDTb) pore with long stem | | Descriptor: | ADP-ribosylating binary toxin binding subunit CdtB, CALCIUM ION, CdtA | | Authors: | Yamada, T, Kawamoto, A, Yoshida, T, Sato, Y, Kato, T, Tsuge, H. | | Deposit date: | 2021-10-11 | | Release date: | 2022-10-26 | | Last modified: | 2024-06-19 | | Method: | ELECTRON MICROSCOPY (2.64 Å) | | Cite: | Cryo-EM structures of the translocational binary toxin complex CDTa-bound CDTb-pore from Clostridioides difficile.
Nat Commun, 13, 2022
|
|
7VNJ
 
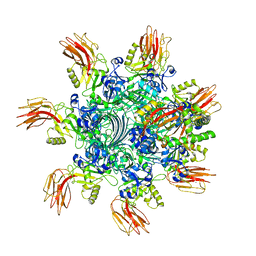 | | Complex structure of Clostridioides difficile enzymatic component (CDTa) and binding component (CDTb) pore with short stem | | Descriptor: | ADP-ribosylating binary toxin binding subunit CdtB, ADP-ribosyltransferase enzymatic component, CALCIUM ION | | Authors: | Yamada, T, Kawamoto, A, Yoshida, T, Sato, Y, Kato, T, Tsuge, H. | | Deposit date: | 2021-10-11 | | Release date: | 2022-10-26 | | Last modified: | 2024-06-19 | | Method: | ELECTRON MICROSCOPY (2.56 Å) | | Cite: | Cryo-EM structures of the translocational binary toxin complex CDTa-bound CDTb-pore from Clostridioides difficile.
Nat Commun, 13, 2022
|
|
8WQP
 
 | |
6JI1
 
 | |
7WZN
 
 | | PSI-LHCI from Chlamydomonas reinhardtii with bound ferredoxin | | Descriptor: | CHLOROPHYLL A, CHLOROPHYLL A ISOMER, CHLOROPHYLL B, ... | | Authors: | Kurisu, G, Gerle, C, Mitsuoka, K, Kawamoto, A, Tanaka, H. | | Deposit date: | 2022-02-18 | | Release date: | 2023-02-22 | | Last modified: | 2024-06-26 | | Method: | ELECTRON MICROSCOPY (4.9 Å) | | Cite: | Three structures of PSI-LHCI from Chlamydomonas reinhardtii suggest a resting state re-activated by ferredoxin.
Biochim Biophys Acta Bioenerg, 1864, 2023
|
|
7Y13
 
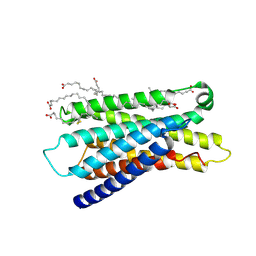 | | Cryo-EM structure of apo-state MrgD-Gi complex (local) | | Descriptor: | PALMITIC ACID, Soluble cytochrome b562,Mas-related G-protein coupled receptor member D | | Authors: | Suzuki, S, Iida, M, Kawamoto, A, Oshima, A. | | Deposit date: | 2022-06-06 | | Release date: | 2022-07-20 | | Last modified: | 2024-10-30 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Structural insight into the activation mechanism of MrgD with heterotrimeric Gi-protein revealed by cryo-EM.
Commun Biol, 5, 2022
|
|
7Y12
 
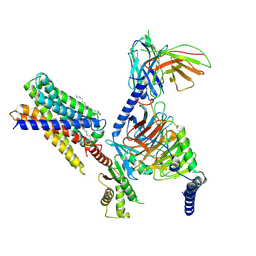 | | Cryo-EM structure of MrgD-Gi complex with beta-alanine | | Descriptor: | BETA-ALANINE, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, ... | | Authors: | Suzuki, S, Iida, M, Kawamoto, A, Oshima, A. | | Deposit date: | 2022-06-06 | | Release date: | 2022-07-20 | | Last modified: | 2024-10-16 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Structural insight into the activation mechanism of MrgD with heterotrimeric Gi-protein revealed by cryo-EM.
Commun Biol, 5, 2022
|
|
7Y14
 
 | | Cryo-EM structure of MrgD-Gi complex with beta-alanine (local) | | Descriptor: | BETA-ALANINE, PALMITIC ACID, Soluble cytochrome b562,Mas-related G-protein coupled receptor member D | | Authors: | Suzuki, S, Iida, M, Kawamoto, A, Oshima, A. | | Deposit date: | 2022-06-06 | | Release date: | 2022-07-20 | | Last modified: | 2024-10-09 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Structural insight into the activation mechanism of MrgD with heterotrimeric Gi-protein revealed by cryo-EM.
Commun Biol, 5, 2022
|
|
7Y15
 
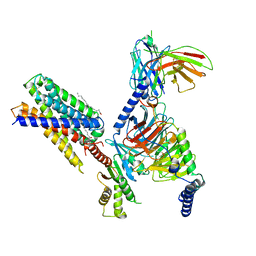 | | Cryo-EM structure of apo-state MrgD-Gi complex | | Descriptor: | Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, Guanine nucleotide-binding protein G(i) subunit alpha-1, ... | | Authors: | Suzuki, S, Iida, M, Kawamoto, A, Oshima, A. | | Deposit date: | 2022-06-06 | | Release date: | 2022-07-20 | | Last modified: | 2022-11-23 | | Method: | ELECTRON MICROSCOPY (2.9 Å) | | Cite: | Structural insight into the activation mechanism of MrgD with heterotrimeric Gi-protein revealed by cryo-EM.
Commun Biol, 5, 2022
|
|
7WYI
 
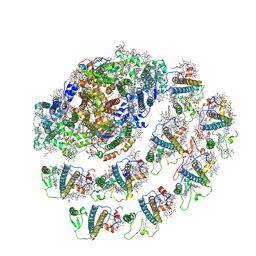 | | Native Photosystem I of Chlamydomonas reinhardtii | | Descriptor: | CHLOROPHYLL A, CHLOROPHYLL A ISOMER, CHLOROPHYLL B, ... | | Authors: | Kurisu, G, Gerle, C, Mitsuoka, K, Kawamoto, A, Tanaka, H. | | Deposit date: | 2022-02-16 | | Release date: | 2023-02-22 | | Last modified: | 2024-06-26 | | Method: | ELECTRON MICROSCOPY (3.9 Å) | | Cite: | Three structures of PSI-LHCI from Chlamydomonas reinhardtii suggest a resting state re-activated by ferredoxin.
Biochim Biophys Acta Bioenerg, 1864, 2023
|
|
6LU1
 
 | | Cyanobacterial PSI Monomer from T. elongatus by Single Particle CRYO-EM at 3.2 A Resolution | | Descriptor: | 1,2-DIPALMITOYL-PHOSPHATIDYL-GLYCEROLE, 1,2-DISTEAROYL-MONOGALACTOSYL-DIGLYCERIDE, BETA-CAROTENE, ... | | Authors: | Kurisu, G, Coruh, O, Tanaka, H, Gerle, C, Kawamoto, A, Kato, T, Namba, K, Nowaczyk, M.M, Rogner, M, Misumi, Y, Frank, A, Eithar, E.M. | | Deposit date: | 2020-01-24 | | Release date: | 2021-03-17 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Cryo-EM structure of a functional monomeric Photosystem I from Thermosynechococcus elongatus reveals red chlorophyll cluster.
Commun Biol, 4, 2021
|
|
7WLM
 
 | | The Cryo-EM structure of siphonaxanthin chlorophyll a/b type light-harvesting complex II | | Descriptor: | (1R,3R)-6-{(3E,5E,7E,9E,11E,13E,15E,17E)-18-[(1S,4R,6R)-4-HYDROXY-2,2,6-TRIMETHYL-7-OXABICYCLO[4.1.0]HEPT-1-YL]-3,7,12,16-TETRAMETHYLOCTADECA-1,3,5,7,9,11,13,15,17-NONAENYLIDENE}-1,5,5-TRIMETHYLCYCLOHEXANE-1,3-DIOL, 1,2-DIPALMITOYL-PHOSPHATIDYL-GLYCEROLE, CHLOROPHYLL A, ... | | Authors: | Seki, S, Nakaniwa, T, Castro-Hartmann, P, Sader, K, Kawamoto, A, Tanaka, H, Qian, P, Kurisu, G, Fujii, R. | | Deposit date: | 2022-01-13 | | Release date: | 2022-11-23 | | Last modified: | 2024-06-26 | | Method: | ELECTRON MICROSCOPY (2.8 Å) | | Cite: | Structural insights into blue-green light utilization by marine green algal light harvesting complex II at 2.78 angstrom.
Bba Adv, 2, 2022
|
|
8WIY
 
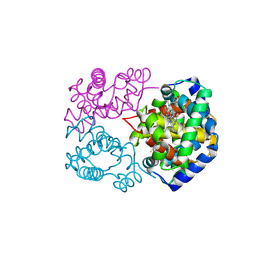 | | cryo-EM structure of alligator haemoglobin in oxy form | | Descriptor: | Hemoglobin subunit alpha, Hemoglobin subunit beta, OXYGEN MOLECULE, ... | | Authors: | Takahashi, K, Lee, Y, Fago, A, Bautista, N.M, Kawamoto, A, Kurisu, G, Storz, J, Nishizawa, T, Tame, J.R.H. | | Deposit date: | 2023-09-25 | | Release date: | 2024-08-14 | | Method: | ELECTRON MICROSCOPY (2.31 Å) | | Cite: | The unique allosteric property of crocodilian haemoglobin elucidated by cryo-EM.
Nat Commun, 15, 2024
|
|
8WIZ
 
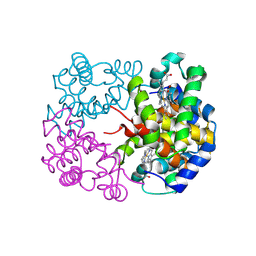 | | cryo-EM structure of alligator haemoglobin in deoxy form | | Descriptor: | BICARBONATE ION, Hemoglobin subunit alpha, Hemoglobin subunit beta, ... | | Authors: | Takahashi, K, Lee, Y, Fago, A, Bautista, N.M, Kawamoto, A, Kurisu, G, Storz, J, Nishizawa, T, Tame, J.R.H. | | Deposit date: | 2023-09-25 | | Release date: | 2024-08-14 | | Method: | ELECTRON MICROSCOPY (2.2 Å) | | Cite: | The unique allosteric property of crocodilian haemoglobin elucidated by cryo-EM.
Nat Commun, 15, 2024
|
|
8WJ0
 
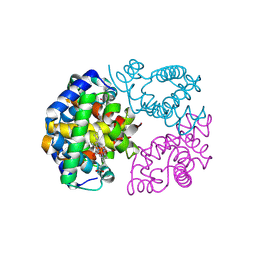 | | cryo-EM structure of human haemoglobin in carbonmonoxy form | | Descriptor: | CARBON MONOXIDE, Hemoglobin subunit alpha, Hemoglobin subunit beta, ... | | Authors: | Takahashi, K, Lee, Y, Fago, A, Bautista, N.M, Kawamoto, A, Kurisu, G, Storz, J, Nishizawa, T, Tame, J.R.H. | | Deposit date: | 2023-09-25 | | Release date: | 2024-08-14 | | Method: | ELECTRON MICROSCOPY (2.24 Å) | | Cite: | The unique allosteric property of crocodilian haemoglobin elucidated by cryo-EM.
Nat Commun, 15, 2024
|
|
8WJ1
 
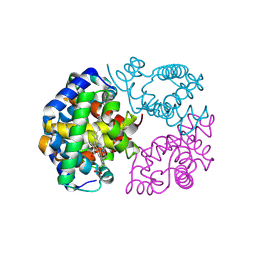 | | cryo-EM structure of human haemoglobin in oxy form | | Descriptor: | Hemoglobin subunit alpha, Hemoglobin subunit beta, OXYGEN MOLECULE, ... | | Authors: | Takahashi, K, Lee, Y, Fago, A, Bautista, N.M, Kawamoto, A, Kurisu, G, Storz, J, Nishizawa, T, Tame, J.R.H. | | Deposit date: | 2023-09-25 | | Release date: | 2024-08-14 | | Method: | ELECTRON MICROSCOPY (2.27 Å) | | Cite: | The unique allosteric property of crocodilian haemoglobin elucidated by cryo-EM.
Nat Commun, 15, 2024
|
|
8WIX
 
 | | cryo-EM structure of alligator haemoglobin in carbonmonoxy form | | Descriptor: | CARBON MONOXIDE, Hemoglobin subunit alpha, Hemoglobin subunit beta, ... | | Authors: | Takahashi, K, Lee, Y, Fago, A, Bautista, N.M, Kawamoto, A, Kurisu, G, Storz, J, Nishizawa, T, Tame, J.R.H. | | Deposit date: | 2023-09-25 | | Release date: | 2024-08-14 | | Method: | ELECTRON MICROSCOPY (2.29 Å) | | Cite: | The unique allosteric property of crocodilian haemoglobin elucidated by cryo-EM.
Nat Commun, 15, 2024
|
|
8WJ2
 
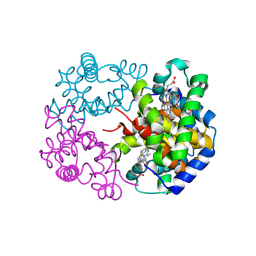 | | cryo-EM structure of human haemoglobin in deoxy form | | Descriptor: | Hemoglobin subunit alpha, Hemoglobin subunit beta, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Takahashi, K, Lee, Y, Fago, A, Bautista, N.M, Kawamoto, A, Kurisu, G, Storz, J, Nishizawa, T, Tame, J.R.H. | | Deposit date: | 2023-09-25 | | Release date: | 2024-08-14 | | Method: | ELECTRON MICROSCOPY (2.35 Å) | | Cite: | The unique allosteric property of crocodilian haemoglobin elucidated by cryo-EM.
Nat Commun, 15, 2024
|
|
6KLX
 
 | | Pore structure of Iota toxin binding component (Ib) | | Descriptor: | CALCIUM ION, Iota toxin component Ib | | Authors: | Yoshida, T, Yamada, T, Kawamoto, A, Mitsuoka, K, Iwasaki, K, Tsuge, H. | | Deposit date: | 2019-07-30 | | Release date: | 2020-01-15 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (2.9 Å) | | Cite: | Cryo-EM structures reveal translocational unfolding in the clostridial binary iota toxin complex.
Nat.Struct.Mol.Biol., 27, 2020
|
|
6KLO
 
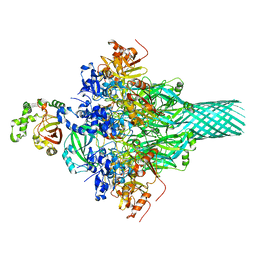 | | Complex structure of Iota toxin enzymatic component (Ia) and binding component (Ib) pore with short stem | | Descriptor: | CALCIUM ION, Iota toxin component Ia, Iota toxin component Ib | | Authors: | Yoshida, T, Yamada, T, Kawamoto, A, Mitsuoka, K, Iwasaki, K, Tsuge, H. | | Deposit date: | 2019-07-30 | | Release date: | 2020-01-15 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (2.8 Å) | | Cite: | Cryo-EM structures reveal translocational unfolding in the clostridial binary iota toxin complex.
Nat.Struct.Mol.Biol., 27, 2020
|
|
6KLW
 
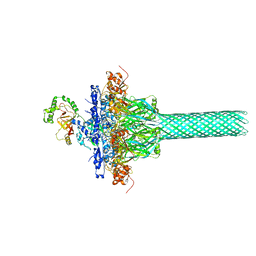 | | Complex structure of Iota toxin enzymatic component (Ia) and binding component (Ib) pore with long stem | | Descriptor: | CALCIUM ION, Iota toxin component Ia, Iota toxin component Ib | | Authors: | Yoshida, T, Yamada, T, Kawamoto, A, Mitsuoka, K, Iwasaki, K, Tsuge, H. | | Deposit date: | 2019-07-30 | | Release date: | 2020-01-15 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (2.9 Å) | | Cite: | Cryo-EM structures reveal translocational unfolding in the clostridial binary iota toxin complex.
Nat.Struct.Mol.Biol., 27, 2020
|
|
