5TXR
 
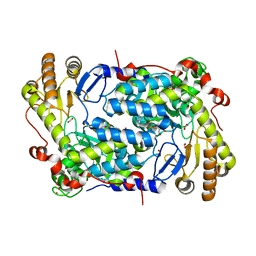 | | Structure of ALAS from S. cerevisiae non-covalently bound to PLP cofactor | | Descriptor: | 5-aminolevulinate synthase, mitochondrial, FORMIC ACID, ... | | Authors: | Brown, B.L, Grant, R.A, Kardon, J.R, Sauer, R.T, Baker, T.A. | | Deposit date: | 2016-11-17 | | Release date: | 2018-03-28 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structure of the Mitochondrial Aminolevulinic Acid Synthase, a Key Heme Biosynthetic Enzyme.
Structure, 26, 2018
|
|
5TXT
 
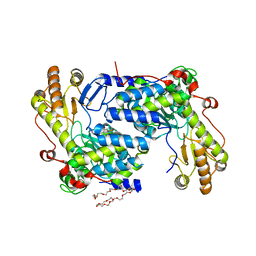 | | Structure of asymmetric apo/holo ALAS dimer from S. cerevisiae | | Descriptor: | 3,6,9,12,15,18,21,24,27,30,33,36,39-TRIDECAOXAHENTETRACONTANE-1,41-DIOL, 5-aminolevulinate synthase, mitochondrial, ... | | Authors: | Brown, B.L, Grant, R.A, Kardon, J.R, Sauer, R.T, Baker, T.A. | | Deposit date: | 2016-11-17 | | Release date: | 2018-03-28 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Structure of the Mitochondrial Aminolevulinic Acid Synthase, a Key Heme Biosynthetic Enzyme.
Structure, 26, 2018
|
|
7M1M
 
 | | Crystal structure of Pseudomonas aeruginosa ClpP1 | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, ATP-dependent Clp protease proteolytic subunit | | Authors: | Mawla, G.D, Grant, R.A, Baker, T.A, Sauer, R.T. | | Deposit date: | 2021-03-13 | | Release date: | 2021-03-24 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | ClpP1P2 peptidase activity promotes biofilm formation in Pseudomonas aeruginosa.
Mol.Microbiol., 115, 2021
|
|
7M1L
 
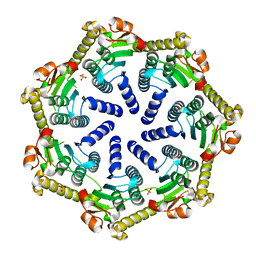 | | Crystal structure of Pseudomonas aeruginosa ClpP2 | | Descriptor: | ATP-dependent Clp protease proteolytic subunit, PHOSPHATE ION | | Authors: | Hall, B.M, Grant, R.A, Baker, T.A, Sauer, R.T. | | Deposit date: | 2021-03-13 | | Release date: | 2021-03-24 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | ClpP1P2 peptidase activity promotes biofilm formation in Pseudomonas aeruginosa.
Mol.Microbiol., 115, 2021
|
|
