1H6N
 
 | | Formation of a tyrosyl radical intermediate in Proteus mirabilis catalase by directed mutagenesis and consequences for nucleotide reactivity | | Descriptor: | ACETATE ION, CATALASE, PROTOPORPHYRIN IX CONTAINING FE, ... | | Authors: | Andreoletti, P, Sainz, G, Jaquinod, M, Gagnon, J, Jouve, H.M. | | Deposit date: | 2001-06-20 | | Release date: | 2003-10-03 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.11 Å) | | Cite: | High Resolution Structure and Biochemical Properties of a Recombinant Proteus Mirabilis Catalase Depleted in Iron.
Proteins: Struct.,Funct., Genet., 50, 2003
|
|
1IMT
 
 | | MAMBA INTESTINAL TOXIN 1, NMR, 39 STRUCTURES | | Descriptor: | INTESTINAL TOXIN 1 | | Authors: | Boisbouvier, J, Albrand, J.-P, Blackledge, M, Jaquinod, M, Schweitz, H, Lazdunski, M, Marion, D. | | Deposit date: | 1998-04-14 | | Release date: | 1999-04-20 | | Last modified: | 2022-02-23 | | Method: | SOLUTION NMR | | Cite: | A structural homologue of colipase in black mamba venom revealed by NMR floating disulphide bridge analysis.
J.Mol.Biol., 283, 1998
|
|
1E93
 
 | | High resolution structure and biochemical properties of a recombinant catalase depleted in iron | | Descriptor: | ACETATE ION, CATALASE, PROTOPORPHYRIN IX CONTAINING FE, ... | | Authors: | Andreoletti, P, Sainz, G, Jaquinod, M, Gagnon, J, Jouve, H.M. | | Deposit date: | 2000-10-05 | | Release date: | 2000-10-13 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | High-Resolution Structure and Biochemical Properties of a Recombinant Proteus Mirabilis Catalase Depleted in Iron.
Proteins: Struct.,Funct., Genet., 50, 2003
|
|
2RGV
 
 | |
3HB6
 
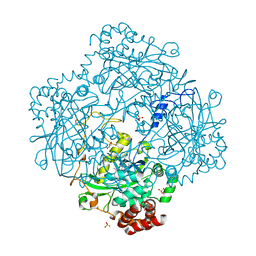 | | Inactive mutant H54F of Proteus mirabilis catalase | | Descriptor: | Catalase, GLYCEROL, PROTOPORPHYRIN IX CONTAINING FE, ... | | Authors: | Andeoletti, P, Jouve, H.M, Gouet, P. | | Deposit date: | 2009-05-04 | | Release date: | 2009-05-12 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Verdoheme formation in Proteus mirabilis catalase.
Biochim.Biophys.Acta, 1790, 2009
|
|
1E67
 
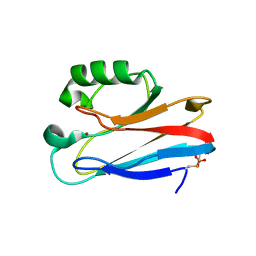 | | Zn-Azurin from Pseudomonas aeruginosa | | Descriptor: | AZURIN, NITRATE ION, ZINC ION | | Authors: | Nar, H, Messerschmidt, A. | | Deposit date: | 2000-08-09 | | Release date: | 2000-08-16 | | Last modified: | 2017-07-12 | | Method: | X-RAY DIFFRACTION (2.14 Å) | | Cite: | Characterization and Crystal Structure of Zinc Azurin, a by-Product of Heterologous Expression in Escherichia Coli of Pseudomonas Aeruginosa Copper Azurin
Eur.J.Biochem., 205, 1992
|
|
1B9P
 
 | |
1B9Q
 
 | |
1OCY
 
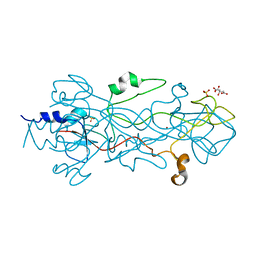 | | Structure of the receptor-binding domain of the bacteriophage T4 short tail fibre | | Descriptor: | BACTERIOPHAGE T4 SHORT TAIL FIBRE, CITRIC ACID, SULFATE ION, ... | | Authors: | Thomassen, E, Gielen, G, Schuetz, M, Miller, S, van Raaij, M.J. | | Deposit date: | 2003-02-11 | | Release date: | 2003-07-24 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | The Structure of the Receptor-Binding Domain of the Bacteriophage T4 Short Tail Fibre Reveals a Knitted Trimeric Metal-Binding Fold
J.Mol.Biol., 331, 2003
|
|
5LYE
 
 | |
2CF4
 
 | | Pyrococcus horikoshii TET1 peptidase can assemble into a tetrahedron or a large octahedral shell | | Descriptor: | COBALT (II) ION, PROTEIN PH0519 | | Authors: | Vellieux, F.M.D, Schoehn, G, Dura, M.A, Roussel, A, Franzetti, B. | | Deposit date: | 2006-02-15 | | Release date: | 2006-09-14 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (3.08 Å) | | Cite: | An Archaeal Peptidase Assembles Into Two Different Quaternary Structures: A Tetrahedron and a Giant Octahedron.
J.Biol.Chem., 281, 2006
|
|
