6LXV
 
 | | Cryo-EM structure of phosphoketolase from Bifidobacterium longum | | Descriptor: | CALCIUM ION, Phosphoketolase, THIAMINE DIPHOSPHATE | | Authors: | Nakata, K, Miyazaki, N, Yamaguchi, H, Hirose, M, Miyano, H, Mizukoshi, T, Kashiwagi, T, Iwasaki, K. | | Deposit date: | 2020-02-12 | | Release date: | 2021-02-17 | | Last modified: | 2024-05-29 | | Method: | ELECTRON MICROSCOPY (2.1 Å) | | Cite: | High-resolution structure of phosphoketolase from Bifidobacterium longum determined by cryo-EM single-particle analysis.
J.Struct.Biol., 214, 2022
|
|
5GJE
 
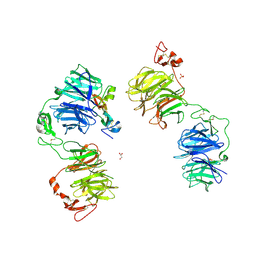 | | Three-dimensional reconstruction of human LRP6 ectodomain complexed with Dkk1 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Matoba, K, Mihara, E, Tamura-Kawakami, K, Hirai, H, Thompson, S, Iwasaki, K, Takagi, J. | | Deposit date: | 2016-06-29 | | Release date: | 2017-01-18 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (21 Å) | | Cite: | Conformational Freedom of the LRP6 Ectodomain Is Regulated by N-glycosylation and the Binding of the Wnt Antagonist Dkk1
Cell Rep, 18, 2017
|
|
6S2C
 
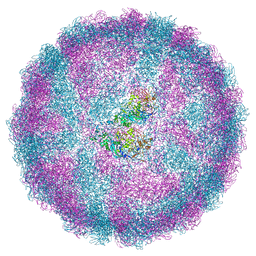 | | Acquired functional capsid structures in metazoan totivirus-like dsRNA virus. | | Descriptor: | Capsid protein | | Authors: | Okamoto, K, Larsson, S.D.D, Maia, R.N.C.F, Murata, K, Hajdu, J, Iwasaki, K, Miyazaki, N. | | Deposit date: | 2019-06-20 | | Release date: | 2020-04-29 | | Last modified: | 2024-05-22 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Acquired Functional Capsid Structures in Metazoan Totivirus-like dsRNA Virus.
Structure, 28, 2020
|
|
8WDV
 
 | | Photosynthetic LH1-RC complex from the purple sulfur bacterium Allochromatium vinosum purified by Ca2+-DEAE | | Descriptor: | (1R)-2-{[{[(2S)-2,3-DIHYDROXYPROPYL]OXY}(HYDROXY)PHOSPHORYL]OXY}-1-[(PALMITOYLOXY)METHYL]ETHYL (11E)-OCTADEC-11-ENOATE, (2S)-3-hydroxypropane-1,2-diyl dihexadecanoate, Antenna complex alpha/beta subunit, ... | | Authors: | Tani, K, Kanno, R, Harada, A, Kobayashi, A, Minamino, A, Nakamura, N, Ji, X.-C, Purba, E.R, Hall, M, Yu, L.-J, Madigan, M.T, Mizoguchi, A, Iwasaki, K, Humbel, B.M, Kimura, Y, Wang-Otomo, Z.-Y. | | Deposit date: | 2023-09-16 | | Release date: | 2024-02-21 | | Last modified: | 2024-05-08 | | Method: | ELECTRON MICROSCOPY (2.24 Å) | | Cite: | High-resolution structure and biochemical properties of the LH1-RC photocomplex from the model purple sulfur bacterium, Allochromatium vinosum.
Commun Biol, 7, 2024
|
|
8WDU
 
 | | Photosynthetic LH1-RC complex from the purple sulfur bacterium Allochromatium vinosum purified by sucrose density | | Descriptor: | (1R)-2-{[{[(2S)-2,3-DIHYDROXYPROPYL]OXY}(HYDROXY)PHOSPHORYL]OXY}-1-[(PALMITOYLOXY)METHYL]ETHYL (11E)-OCTADEC-11-ENOATE, (2S)-3-hydroxypropane-1,2-diyl dihexadecanoate, Antenna complex alpha/beta subunit, ... | | Authors: | Tani, K, Kanno, R, Harada, A, Kobayashi, A, Minamino, A, Nakamura, N, Ji, X.-C, Purba, E.R, Hall, M, Yu, L.-J, Madigan, M.T, Mizoguchi, A, Iwasaki, K, Humbel, B.M, Kimura, Y, Wang-Otomo, Z.-Y. | | Deposit date: | 2023-09-16 | | Release date: | 2024-02-21 | | Last modified: | 2024-05-08 | | Method: | ELECTRON MICROSCOPY (2.24 Å) | | Cite: | High-resolution structure and biochemical properties of the LH1-RC photocomplex from the model purple sulfur bacterium, Allochromatium vinosum.
Commun Biol, 7, 2024
|
|
6RVV
 
 | | Structure of left-handed protein cage consisting of 24 eleven-membered ring proteins held together by gold (I) bridges. | | Descriptor: | GOLD ION, Transcription attenuation protein MtrB | | Authors: | Malay, A.D, Miyazaki, N, Biela, A.P, Iwasaki, K, Heddle, J.G. | | Deposit date: | 2019-06-03 | | Release date: | 2019-06-12 | | Last modified: | 2024-05-15 | | Method: | ELECTRON MICROSCOPY (3.7 Å) | | Cite: | An ultra-stable gold-coordinated protein cage displaying reversible assembly.
Nature, 569, 2019
|
|
6RVW
 
 | | Structure of right-handed protein cage consisting of 24 eleven-membered ring proteins held together by gold (I) bridges. | | Descriptor: | GOLD ION, Transcription attenuation protein MtrB | | Authors: | Malay, A.D, Miyazaki, N, Biela, A.P, Iwasaki, K, Heddle, J.G. | | Deposit date: | 2019-06-03 | | Release date: | 2019-06-12 | | Last modified: | 2024-05-15 | | Method: | ELECTRON MICROSCOPY (3.7 Å) | | Cite: | An ultra-stable gold-coordinated protein cage displaying reversible assembly.
Nature, 569, 2019
|
|
7WU9
 
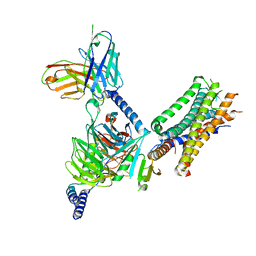 | | Cryo-EM structure of the human EP3-Gi signaling complex | | Descriptor: | Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, Guanine nucleotide-binding protein G(i) subunit alpha-1, ... | | Authors: | Suno, R, Sugita, Y, Morimoto, K, Iwasaki, K, Kato, T, Kobayashi, T. | | Deposit date: | 2022-02-07 | | Release date: | 2022-08-17 | | Last modified: | 2023-10-04 | | Method: | ELECTRON MICROSCOPY (3.375 Å) | | Cite: | Structural insights into the G protein selectivity revealed by the human EP3-G i signaling complex.
Cell Rep, 40, 2022
|
|
7X30
 
 | | Capsid structure of Staphylococcus jumbo bacteriophage S6 | | Descriptor: | Hoc-like protein ORF90, Major structural protein ORF12 | | Authors: | Koibuchi, W, Uchiyama, J, Matsuzaki, S, Murata, K, Iwasaki, K, Miyazaki, N. | | Deposit date: | 2022-02-27 | | Release date: | 2023-12-06 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | Capsid structure of Staphylococcus jumbo bacteriophage S6
To Be Published
|
|
7WMP
 
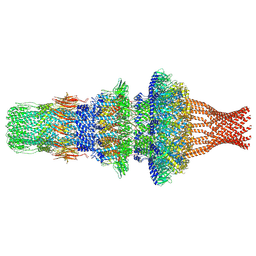 | | Tail structure of Helicobacter pylori bacteriophage KHP30 | | Descriptor: | Adaptor protein gp12, Nozzle protein gp25, Portal protein | | Authors: | Kamiya, R, Uchiyama, J, Matsuzaki, S, Murata, K, Iwasaki, K, Miyazaki, N. | | Deposit date: | 2022-01-15 | | Release date: | 2023-03-01 | | Last modified: | 2024-06-26 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | Cryo-EM structure of Helicobacter pylori bacteriophage KHP30
To Be Published
|
|
1IQ4
 
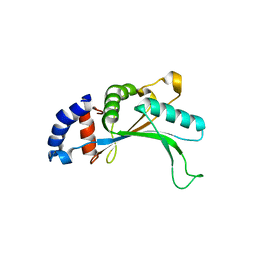 | | 5S-RRNA BINDING RIBOSOMAL PROTEIN L5 FROM BACILLUS STEAROTHERMOPHILUS | | Descriptor: | 50S RIBOSOMAL PROTEIN L5 | | Authors: | Nakashima, T, Yao, M, Kawamura, S, Iwasaki, K, Kimura, M, Tanaka, I. | | Deposit date: | 2001-06-13 | | Release date: | 2001-06-27 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Ribosomal protein L5 has a highly twisted concave surface and flexible arms responsible for rRNA binding.
RNA, 7, 2001
|
|
7DOD
 
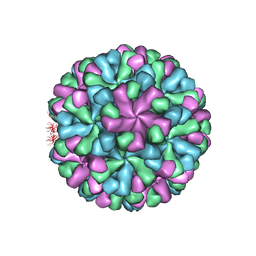 | | Capsid structure of human sapovirus | | Descriptor: | Calicivirin | | Authors: | Miyazaki, N, Murakami, K, Oka, T, Iwasaki, K, Katayama, K, Murata, K. | | Deposit date: | 2020-12-14 | | Release date: | 2021-12-15 | | Last modified: | 2024-06-05 | | Method: | ELECTRON MICROSCOPY (2.9 Å) | | Cite: | Atomic structure of human sapovirus capsid by single particle cryo-electron microscopy
To Be Published
|
|
7DOU
 
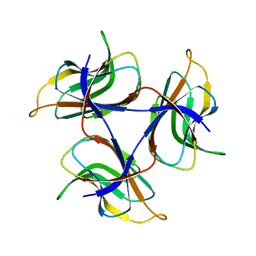 | | Trimeric cement protein structure of Helicobacter pylori bacteriophage KHP40 | | Descriptor: | Cement protein gp16 | | Authors: | Kamiya, R, Uchiyama, J, Matsuzaki, S, Murata, K, Iwasaki, K, Miyazaki, N. | | Deposit date: | 2020-12-17 | | Release date: | 2021-10-27 | | Last modified: | 2024-06-05 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Acid-stable capsid structure of Helicobacter pylori bacteriophage KHP30 by single-particle cryoelectron microscopy.
Structure, 30, 2022
|
|
7DN2
 
 | | Acidic stable capsid structure of Helicobacter pylori bacteriophage KHP30 | | Descriptor: | Cement protein gp15, Major structural protein ORF14 | | Authors: | Kamiya, R, Uchiyama, J, Matsuzaki, S, Murata, K, Iwasaki, K, Miyazaki, N. | | Deposit date: | 2020-12-08 | | Release date: | 2021-10-27 | | Last modified: | 2024-05-29 | | Method: | ELECTRON MICROSCOPY (2.7 Å) | | Cite: | Acid-stable capsid structure of Helicobacter pylori bacteriophage KHP30 by single-particle cryoelectron microscopy.
Structure, 30, 2022
|
|
7F2P
 
 | | The head structure of Helicobacter pylori bacteriophage KHP40 | | Descriptor: | Cement protein gp16, KHP40 MCP | | Authors: | Kamiya, R, Uchiyama, J, Matsuzaki, S, Murata, K, Iwasaki, K, Miyazaki, N. | | Deposit date: | 2021-06-13 | | Release date: | 2021-10-27 | | Last modified: | 2024-06-12 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Acid-stable capsid structure of Helicobacter pylori bacteriophage KHP30 by single-particle cryoelectron microscopy.
Structure, 30, 2022
|
|
6IUK
 
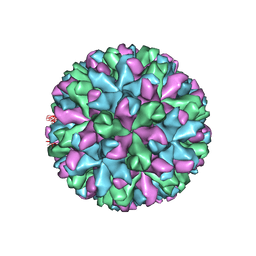 | | Cryo-EM structure of Murine Norovirus capsid | | Descriptor: | Major capsid protein VP1 | | Authors: | Song, C, Miyazaki, N, Iwasaki, K, Katayama, K, Murata, K. | | Deposit date: | 2018-11-28 | | Release date: | 2020-02-26 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | Dynamic rotation of the protruding domain enhances the infectivity of norovirus.
Plos Pathog., 16, 2020
|
|
6KNC
 
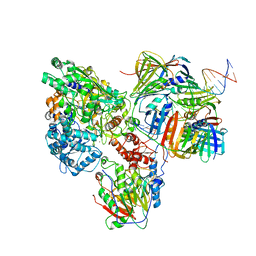 | | PolD-PCNA-DNA (form B) | | Descriptor: | DNA polymerase D DP2 (DNA polymerase II large) subunit, DNA polymerase II small subunit, DNA polymerase sliding clamp 1, ... | | Authors: | Mayanagi, K, Oki, K, Miyazaki, N, Ishino, S, Yamagami, T, Iwasaki, K, Kohda, D, Morikawa, K, Shirai, T, Ishino, Y. | | Deposit date: | 2019-08-05 | | Release date: | 2020-08-05 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (9.3 Å) | | Cite: | Two conformations of DNA polymerase D-PCNA-DNA, an archaeal replisome complex, revealed by cryo-electron microscopy.
Bmc Biol., 18, 2020
|
|
6KNB
 
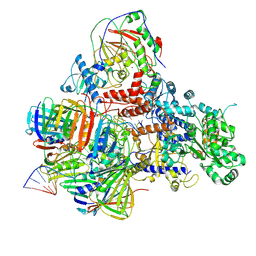 | | PolD-PCNA-DNA (form A) | | Descriptor: | DNA polymerase D DP2 (DNA polymerase II large) subunit, DNA polymerase II small subunit, DNA polymerase sliding clamp 1, ... | | Authors: | Mayanagi, K, Oki, K, Miyazaki, N, Ishino, S, Yamagami, T, Iwasaki, K, Kohda, D, Morikawa, K, Shirai, T, Ishino, Y. | | Deposit date: | 2019-08-05 | | Release date: | 2020-08-05 | | Last modified: | 2021-02-17 | | Method: | ELECTRON MICROSCOPY (6.9 Å) | | Cite: | Two conformations of DNA polymerase D-PCNA-DNA, an archaeal replisome complex, revealed by cryo-electron microscopy.
Bmc Biol., 18, 2020
|
|
6KLX
 
 | | Pore structure of Iota toxin binding component (Ib) | | Descriptor: | CALCIUM ION, Iota toxin component Ib | | Authors: | Yoshida, T, Yamada, T, Kawamoto, A, Mitsuoka, K, Iwasaki, K, Tsuge, H. | | Deposit date: | 2019-07-30 | | Release date: | 2020-01-15 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (2.9 Å) | | Cite: | Cryo-EM structures reveal translocational unfolding in the clostridial binary iota toxin complex.
Nat.Struct.Mol.Biol., 27, 2020
|
|
6KLO
 
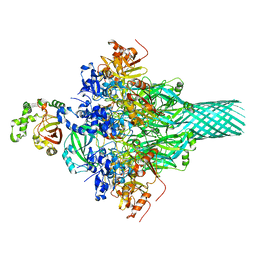 | | Complex structure of Iota toxin enzymatic component (Ia) and binding component (Ib) pore with short stem | | Descriptor: | CALCIUM ION, Iota toxin component Ia, Iota toxin component Ib | | Authors: | Yoshida, T, Yamada, T, Kawamoto, A, Mitsuoka, K, Iwasaki, K, Tsuge, H. | | Deposit date: | 2019-07-30 | | Release date: | 2020-01-15 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (2.8 Å) | | Cite: | Cryo-EM structures reveal translocational unfolding in the clostridial binary iota toxin complex.
Nat.Struct.Mol.Biol., 27, 2020
|
|
6KLW
 
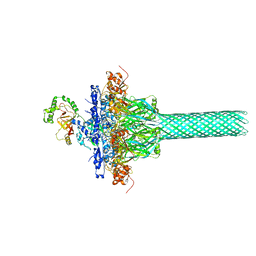 | | Complex structure of Iota toxin enzymatic component (Ia) and binding component (Ib) pore with long stem | | Descriptor: | CALCIUM ION, Iota toxin component Ia, Iota toxin component Ib | | Authors: | Yoshida, T, Yamada, T, Kawamoto, A, Mitsuoka, K, Iwasaki, K, Tsuge, H. | | Deposit date: | 2019-07-30 | | Release date: | 2020-01-15 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (2.9 Å) | | Cite: | Cryo-EM structures reveal translocational unfolding in the clostridial binary iota toxin complex.
Nat.Struct.Mol.Biol., 27, 2020
|
|
3VKF
 
 | | Crystal Structure of Neurexin 1beta/Neuroligin 1 complex | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, Neurexin-1-beta, ... | | Authors: | Tanaka, H, Miyazaki, N, Nogi, T, Iwasaki, K, Takagi, J. | | Deposit date: | 2011-11-15 | | Release date: | 2012-08-01 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (3.3 Å) | | Cite: | Higher-order architecture of cell adhesion mediated by polymorphic synaptic adhesion molecules neurexin and neuroligin.
Cell Rep, 2, 2012
|
|
6AB6
 
 | | Cryo-EM structure of T=3 Penaeus vannamei nodavirus | | Descriptor: | CALCIUM ION, Capsid protein | | Authors: | Chen, N.C, Miyazaki, N, Yoshimura, M, Guan, H.H, Lin, C.C, Iwasaki, K, Chen, C.J. | | Deposit date: | 2018-07-20 | | Release date: | 2019-03-20 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | The atomic structures of shrimp nodaviruses reveal new dimeric spike structures and particle polymorphism.
Commun Biol, 2, 2019
|
|
6AB5
 
 | | Cryo-EM structure of T=1 Penaeus vannamei nodavirus | | Descriptor: | Capsid protein | | Authors: | Chen, N.C, Miyazaki, N, Yoshimura, M, Guan, H.H, Lin, C.C, Iwasaki, K, Chen, C.J. | | Deposit date: | 2018-07-20 | | Release date: | 2019-03-20 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (3.7 Å) | | Cite: | The atomic structures of shrimp nodaviruses reveal new dimeric spike structures and particle polymorphism.
Commun Biol, 2, 2019
|
|
4V60
 
 | | The structure of rat liver vault at 3.5 angstrom resolution | | Descriptor: | Major vault protein | | Authors: | Kato, K, Zhou, Y, Tanaka, H, Yao, M, Yamashita, E, Yoshimura, M, Tsukihara, T. | | Deposit date: | 2008-10-24 | | Release date: | 2014-07-09 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (3.5 Å) | | Cite: | The structure of rat liver vault at 3.5 angstrom resolution
Science, 323, 2009
|
|
