3ONS
 
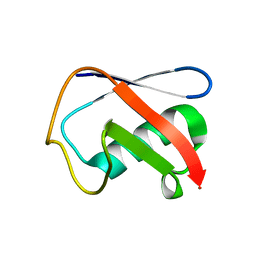 | | Crystal structure of Human Ubiquitin in a new crystal form | | Descriptor: | Ubiquitin | | Authors: | Amodeo, G.A, Huang, K.Y, Mcdermott, A.E, Tong, L. | | Deposit date: | 2010-08-30 | | Release date: | 2011-02-23 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | The structure of human ubiquitin in 2-methyl-2,4-pentanediol: a new conformational switch.
Protein Sci., 20, 2011
|
|
8HZM
 
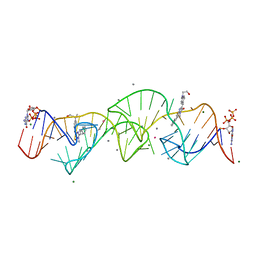 | | A new fluorescent RNA aptamer bound with N, manganese soak | | Descriptor: | (5~{Z})-5-[[4-[2-hydroxyethyl(methyl)amino]phenyl]methylidene]-3-methyl-2-[(~{E})-2-phenylethenyl]imidazol-4-one, GUANOSINE-5'-TRIPHOSPHATE, MAGNESIUM ION, ... | | Authors: | Huang, K.Y, Ren, A.M. | | Deposit date: | 2023-01-09 | | Release date: | 2024-06-19 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structural basis of a small monomeric Clivia fluorogenic RNA with a large Stokes shift.
Nat.Chem.Biol., 2024
|
|
8HZE
 
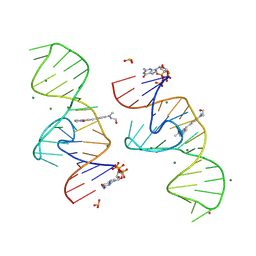 | | A new fluorescent RNA aptamer bound with N | | Descriptor: | (5~{Z})-5-[[4-[2-hydroxyethyl(methyl)amino]phenyl]methylidene]-3-methyl-2-[(~{E})-2-phenylethenyl]imidazol-4-one, GUANOSINE-5'-TRIPHOSPHATE, MAGNESIUM ION, ... | | Authors: | Huang, K.Y, Ren, A.M. | | Deposit date: | 2023-01-09 | | Release date: | 2024-06-19 | | Method: | X-RAY DIFFRACTION (1.59 Å) | | Cite: | Structural basis of a small monomeric Clivia fluorogenic RNA with a large Stokes shift.
Nat.Chem.Biol., 2024
|
|
8HZJ
 
 | | A new fluorescent RNA aptamer bound with N571 | | Descriptor: | (5~{Z})-5-[[3,5-bis(fluoranyl)-4-oxidanyl-phenyl]methylidene]-3-methyl-2-[(~{E})-2-phenylethenyl]imidazol-4-one, GUANOSINE-5'-TRIPHOSPHATE, MAGNESIUM ION, ... | | Authors: | Huang, K.Y, Ren, A.M. | | Deposit date: | 2023-01-09 | | Release date: | 2024-06-19 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structural basis of a small monomeric Clivia fluorogenic RNA with a large Stokes shift.
Nat.Chem.Biol., 2024
|
|
8HZK
 
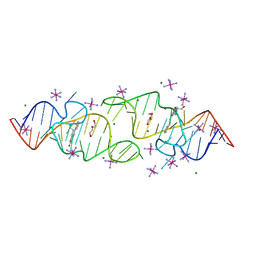 | | A new fluorescent RNA aptamer bound with N, iridium hexammine soak | | Descriptor: | (5~{Z})-5-[[4-[2-hydroxyethyl(methyl)amino]phenyl]methylidene]-3-methyl-2-[(~{E})-2-phenylethenyl]imidazol-4-one, DI(HYDROXYETHYL)ETHER, IRIDIUM HEXAMMINE ION, ... | | Authors: | Huang, K.Y, Ren, A.M. | | Deposit date: | 2023-01-09 | | Release date: | 2024-06-19 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural basis of a small monomeric Clivia fluorogenic RNA with a large Stokes shift.
Nat.Chem.Biol., 2024
|
|
8HZF
 
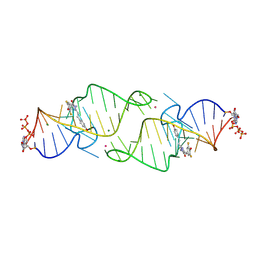 | | A new fluorescent RNA aptamer bound with N565 | | Descriptor: | (5~{Z})-5-[[3,5-bis(fluoranyl)-4-oxidanyl-phenyl]methylidene]-2-[(~{E})-2-(4-hydroxyphenyl)ethenyl]-3-methyl-imidazol-4-one, GUANOSINE-5'-TRIPHOSPHATE, MAGNESIUM ION, ... | | Authors: | Huang, K.Y, Ren, A.M. | | Deposit date: | 2023-01-09 | | Release date: | 2024-06-19 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Structural basis of a small monomeric Clivia fluorogenic RNA with a large Stokes shift.
Nat.Chem.Biol., 2024
|
|
8HZL
 
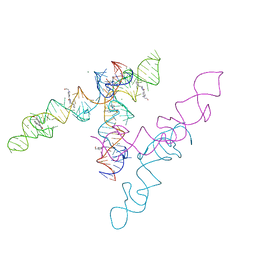 | | A new fluorescent RNA aptamer_III bound with N | | Descriptor: | (5~{Z})-5-[[4-[2-hydroxyethyl(methyl)amino]phenyl]methylidene]-3-methyl-2-[(~{E})-2-phenylethenyl]imidazol-4-one, MAGNESIUM ION, RNA (84-MER) | | Authors: | Huang, K.Y, Ren, A.M. | | Deposit date: | 2023-01-09 | | Release date: | 2024-06-19 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structural basis of a small monomeric Clivia fluorogenic RNA with a large Stokes shift.
Nat.Chem.Biol., 2024
|
|
7EOK
 
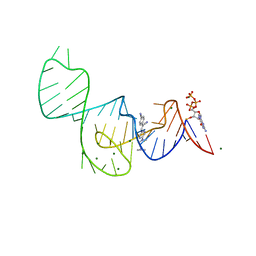 | | Crystal structure of the Pepper aptamer in complex with HBC485 | | Descriptor: | 4-[(~{Z})-1-cyano-2-[5-[2-(dimethylamino)ethyl-methyl-amino]pyrazin-2-yl]ethenyl]benzenecarbonitrile, GUANOSINE-5'-TRIPHOSPHATE, MAGNESIUM ION, ... | | Authors: | Huang, K.Y, Ren, A.M. | | Deposit date: | 2021-04-22 | | Release date: | 2021-11-24 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Structure-based investigation of fluorogenic Pepper aptamer.
Nat.Chem.Biol., 17, 2021
|
|
7EOO
 
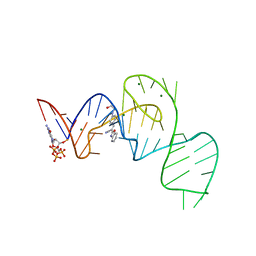 | | Crystal structure of the Pepper aptamer in complex with HBC525 | | Descriptor: | (~{E})-2-(1,3-benzoxazol-2-yl)-3-[4-[2-hydroxyethyl(methyl)amino]phenyl]prop-2-enenitrile, GUANOSINE-5'-TRIPHOSPHATE, MAGNESIUM ION, ... | | Authors: | Huang, K.Y, Ren, A.M. | | Deposit date: | 2021-04-22 | | Release date: | 2021-11-24 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.23 Å) | | Cite: | Structure-based investigation of fluorogenic Pepper aptamer.
Nat.Chem.Biol., 17, 2021
|
|
7EOI
 
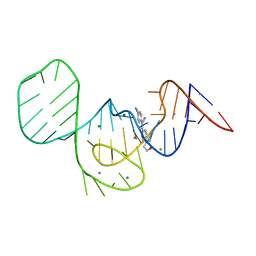 | |
7EOP
 
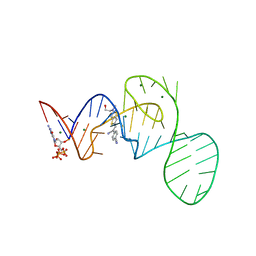 | | Crystal structure of the Pepper aptamer in complex with HBC620 | | Descriptor: | 4-[(~{Z})-1-cyano-2-[5-[2-hydroxyethyl(methyl)amino]thieno[3,2-b]thiophen-2-yl]ethenyl]benzenecarbonitrile, GUANOSINE-5'-TRIPHOSPHATE, MAGNESIUM ION, ... | | Authors: | Huang, K.Y, Ren, A.M. | | Deposit date: | 2021-04-22 | | Release date: | 2021-11-24 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structure-based investigation of fluorogenic Pepper aptamer.
Nat.Chem.Biol., 17, 2021
|
|
7EON
 
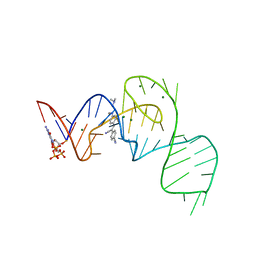 | | Crystal structure of the Pepper aptamer in complex with HBC514 | | Descriptor: | 4-[(~{Z})-1-cyano-2-[4-[2-(dimethylamino)ethyl-methyl-amino]phenyl]ethenyl]benzenecarbonitrile, GUANOSINE-5'-TRIPHOSPHATE, MAGNESIUM ION, ... | | Authors: | Huang, K.Y, Ren, A.M. | | Deposit date: | 2021-04-22 | | Release date: | 2021-11-24 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structure-based investigation of fluorogenic Pepper aptamer.
Nat.Chem.Biol., 17, 2021
|
|
7EOG
 
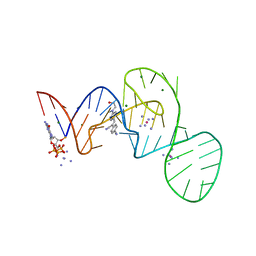 | |
7EOL
 
 | | Crystal structure of the Pepper aptamer in complex with HBC497 | | Descriptor: | 4-[(~{Z})-1-cyano-2-[5-[2-hydroxyethyl(methyl)amino]pyrazin-2-yl]ethenyl]benzenecarbonitrile, GUANOSINE-5'-TRIPHOSPHATE, MAGNESIUM ION, ... | | Authors: | Huang, K.Y, Ren, A.M. | | Deposit date: | 2021-04-22 | | Release date: | 2021-11-24 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.309 Å) | | Cite: | Structure-based investigation of fluorogenic Pepper aptamer.
Nat.Chem.Biol., 17, 2021
|
|
7EOJ
 
 | |
7EOM
 
 | | Crystal structure of the Pepper aptamer in complex with HBC508 | | Descriptor: | 4-[(~{Z})-1-cyano-2-[6-[2-hydroxyethyl(methyl)amino]pyridin-3-yl]ethenyl]benzenecarbonitrile, GUANOSINE-5'-TRIPHOSPHATE, MAGNESIUM ION, ... | | Authors: | Huang, K.Y, Ren, A.M. | | Deposit date: | 2021-04-22 | | Release date: | 2021-11-24 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.703 Å) | | Cite: | Structure-based investigation of fluorogenic Pepper aptamer.
Nat.Chem.Biol., 17, 2021
|
|
7EOH
 
 | | Crystal structure of the Pepper aptamer in complex with HBC | | Descriptor: | 4-[(~{Z})-1-cyano-2-[4-[2-hydroxyethyl(methyl)amino]phenyl]ethenyl]benzenecarbonitrile, MAGNESIUM ION, Pepper (49-MER) | | Authors: | Huang, K.Y, Ren, A.M. | | Deposit date: | 2021-04-22 | | Release date: | 2021-11-24 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.637 Å) | | Cite: | Structure-based investigation of fluorogenic Pepper aptamer.
Nat.Chem.Biol., 17, 2021
|
|
7QU8
 
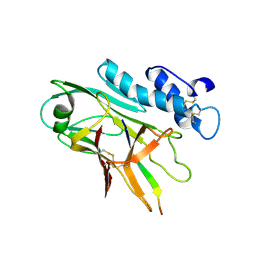 | | ADGRG3/GPR97 Extracellular Region | | Descriptor: | Adhesion G protein-coupled receptor G3 | | Authors: | Zheng-Gerard, C, Chu, T.Y, El Omari, K, Lin, H.H, Seiradake, E. | | Deposit date: | 2022-01-17 | | Release date: | 2022-09-28 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (3.37 Å) | | Cite: | GPR97-mediated PAR2 transactivation via a mPR3-associated macromolecular complex induces inflammatory activation of human neutrophils
Nat Commun, 2022
|
|
6JXG
 
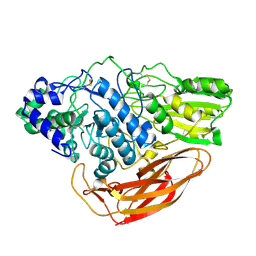 | | Crystasl Structure of Beta-glucosidase D2-BGL from Chaetomella Raphigera | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Beta-glucosidase, alpha-D-mannopyranose | | Authors: | Wang, A.H.-J, Lee, C.C, Kao, M.R, Ho, T.H.D. | | Deposit date: | 2019-04-23 | | Release date: | 2019-11-20 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Chaetomella raphigerabeta-glucosidase D2-BGL has intriguing structural features and a high substrate affinity that renders it an efficient cellulase supplement for lignocellulosic biomass hydrolysis.
Biotechnol Biofuels, 12, 2019
|
|
7QNY
 
 | | The receptor binding domain of SARS-CoV-2 spike glycoprotein in complex with COVOX-58 and COVOX-158 Fabs | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, COVOX-158 heavy chain, COVOX-158 light chain, ... | | Authors: | Zhou, D, Ren, J, Stuart, D.I. | | Deposit date: | 2021-12-23 | | Release date: | 2022-01-19 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.84 Å) | | Cite: | SARS-CoV-2 Omicron-B.1.1.529 leads to widespread escape from neutralizing antibody responses.
Cell, 185, 2022
|
|
7QNW
 
 | | The receptor binding domain of SARS-CoV-2 Omicron variant spike glycoprotein in complex with Beta-55 and EY6A Fabs | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ACETATE ION, Beta-55 heavy chain, ... | | Authors: | Zhou, D, Ren, J, Stuart, D.I. | | Deposit date: | 2021-12-23 | | Release date: | 2022-01-19 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | SARS-CoV-2 Omicron-B.1.1.529 leads to widespread escape from neutralizing antibody responses.
Cell, 185, 2022
|
|
7QNX
 
 | | The receptor binding domain of SARS-CoV-2 spike glycoprotein in complex with Beta-55 and EY6A Fabs | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Beta-55 heavy chain, Beta-55 light chain, ... | | Authors: | Zhou, D, Ren, J, Stuart, D.I. | | Deposit date: | 2021-12-23 | | Release date: | 2022-01-19 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.92 Å) | | Cite: | SARS-CoV-2 Omicron-B.1.1.529 leads to widespread escape from neutralizing antibody responses.
Cell, 185, 2022
|
|
