1V18
 
 | |
1T08
 
 | | Crystal structure of beta-catenin/ICAT helical domain/unphosphorylated APC R3 | | Descriptor: | Adenomatous polyposis coli protein, Beta-catenin, Beta-catenin-interacting protein 1 | | Authors: | Ha, N.-C, Tonozuka, T, Stamos, J.L, Weis, W.I. | | Deposit date: | 2004-04-07 | | Release date: | 2004-10-12 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Mechanism of phosphorylation-dependent binding of APC to beta-catenin and its role in beta-catenin degradation
Mol.Cell, 15, 2004
|
|
1E9Z
 
 | |
1E9Y
 
 | | Crystal structure of Helicobacter pylori urease in complex with acetohydroxamic acid | | Descriptor: | ACETOHYDROXAMIC ACID, NICKEL (II) ION, UREASE SUBUNIT ALPHA, ... | | Authors: | Ha, N.-C, Oh, S.-T, Oh, B.-H. | | Deposit date: | 2000-11-01 | | Release date: | 2001-11-01 | | Last modified: | 2025-04-09 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Supramolecular Assembly and Acid Resistance of Helicobacter Pylori Urease
Nat.Struct.Biol., 8, 2001
|
|
1E3V
 
 | | Crystal structure of ketosteroid isomerase from Psedomonas putida complexed with deoxycholate | | Descriptor: | (3ALPHA,5BETA,12ALPHA)-3,12-DIHYDROXYCHOLAN-24-OIC ACID, STEROID DELTA-ISOMERASE | | Authors: | Ha, N.-C, Kim, M.-S, Kim, J.-S, Oh, B.-H. | | Deposit date: | 2000-06-24 | | Release date: | 2001-03-12 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Detection of Large Pka Perturbations of an Inhibitor and a Catalytic Group at an Enzyme Active Site, a Mechanistic Basis for Catalytic Power of Many Enzymes
J.Biol.Chem., 275, 2000
|
|
1E3R
 
 | | Crystal structure of ketosteroid isomerase mutant D40N (D38N TI numbering) from Pseudomonas putida complexed with androsten-3beta-ol-17-one | | Descriptor: | 3-BETA-HYDROXY-5-ANDROSTEN-17-ONE, ISOMERASE | | Authors: | Ha, N.-C, Kim, M.-S, Hyun, B.-H, Oh, B.-H. | | Deposit date: | 2000-06-22 | | Release date: | 2001-03-05 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Detection of Large Pka Perturbations of an Inhibitor and a Catalytic Group at an Enzyme Active Site, a Mechanistic Basis for Catalytic Power of Many Enzymes
J.Biol.Chem., 275, 2000
|
|
1OGX
 
 | | High Resolution Crystal Structure Of Ketosteroid Isomerase Mutant D40N(D38N, Ti Numbering) from Pseudomonas putida Complexed With Equilenin At 2.0 A Resolution. | | Descriptor: | EQUILENIN, STEROID DELTA-ISOMERASE | | Authors: | Ha, N.-C, Kim, M.-S, Oh, B.-H. | | Deposit date: | 2003-05-17 | | Release date: | 2003-05-20 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Detection of Large Pka Perturbation of an Inhibitor and a Catalytic Group at an Enzyme Active Site, a Mechanistic Basis for Catalytic Power of Many Enzymes
J.Biol.Chem., 275, 2000
|
|
1QLG
 
 | | Crystal structure of phytase with magnesium from Bacillus amyloliquefaciens | | Descriptor: | 3-PHYTASE, CALCIUM ION, MAGNESIUM ION | | Authors: | Shin, S, Ha, N.-C, Oh, B.-H. | | Deposit date: | 1999-08-31 | | Release date: | 2000-02-03 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal Structures of a Novel, Thermostable Phytase in Partially and Fully Calcium-Loaded States
Nat.Struct.Biol., 7, 2000
|
|
3LWG
 
 | |
3LW3
 
 | |
1E97
 
 | |
7D62
 
 | |
6KUI
 
 | |
6KWW
 
 | | HslU from Staphylococcus aureus | | Descriptor: | ATP-dependent protease ATPase subunit HslU | | Authors: | Ha, N.-C, Jeong, S. | | Deposit date: | 2019-09-09 | | Release date: | 2020-07-15 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Cleavage-Dependent Activation of ATP-Dependent Protease HslUV from Staphylococcus aureus .
Mol.Cells, 43, 2020
|
|
6KR1
 
 | | ATP dependent protease HslV from Staphylococcus aureus | | Descriptor: | ATP-dependent protease subunit HslV, SULFATE ION | | Authors: | Ha, N.-C, Jeong, S. | | Deposit date: | 2019-08-20 | | Release date: | 2020-07-15 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Cleavage-Dependent Activation of ATP-Dependent Protease HslUV from Staphylococcus aureus .
Mol.Cells, 43, 2020
|
|
6KHQ
 
 | |
6KGZ
 
 | | bacterial cystathionine gamma-lyase MccB of Staphylococcus aureus | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, Cystathionine gamma-lyase | | Authors: | Ha, N.-C, Lee, D. | | Deposit date: | 2019-07-12 | | Release date: | 2020-03-25 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal Structure of Bacterial Cystathionine Gamma-Lyase in The Cysteine Biosynthesis Pathway of Staphylococcus aureus
Crystals, 9, 2019
|
|
5FHK
 
 | | Regulatory domain of AphB in Vibrio vulnificus | | Descriptor: | LysR family transcriptional regulator | | Authors: | Song, S, Ha, N.-C. | | Deposit date: | 2015-12-22 | | Release date: | 2017-01-11 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.905 Å) | | Cite: | Crystal Structure of the Regulatory Domain of AphB from Vibrio vulnificus, a Virulence Gene Regulator
Mol. Cells, 40, 2017
|
|
3FPP
 
 | | Crystal structure of E.coli MacA | | Descriptor: | Macrolide-specific efflux protein macA | | Authors: | Yum, S, Xu, Y, Piao, S, Ha, N.-C. | | Deposit date: | 2009-01-06 | | Release date: | 2009-04-28 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.99 Å) | | Cite: | Crystal structure of the periplasmic component of a tripartite macrolide-specific efflux pump
J.Mol.Biol., 387, 2009
|
|
5HFG
 
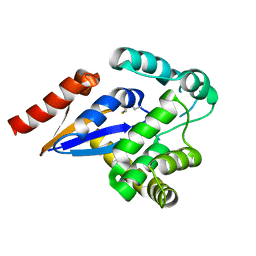 | | Cytosolic disulfide reductase DsbM from Pseudomonas aeruginosa | | Descriptor: | Uncharacterized protein, cytosolic disulfide reductase DsbM | | Authors: | Jo, I, Ha, N.-C. | | Deposit date: | 2016-01-07 | | Release date: | 2016-10-26 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.823 Å) | | Cite: | Crystal structures of the disulfide reductase DsbM from Pseudomonas aeruginosa
Acta Crystallogr D Struct Biol, 72, 2016
|
|
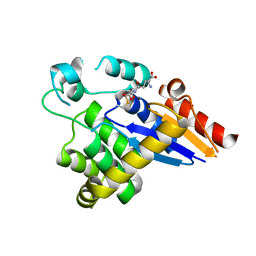
5HFI
 
 | |
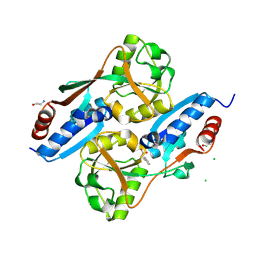
7XRO
 
 | |
7X5D
 
 | |
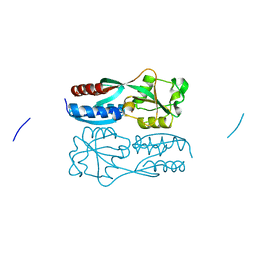
5B7D
 
 | |
5B70
 
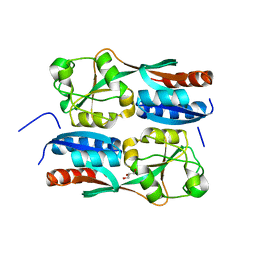 | | OxyR2 E204G regulatory domain from Vibrio vulnificus | | Descriptor: | GLYCEROL, LysR family transcriptional regulator | | Authors: | Jo, I, Ha, N.-C. | | Deposit date: | 2016-06-02 | | Release date: | 2017-03-15 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | The hydrogen peroxide hypersensitivity of OxyR2 in Vibrio vulnificus depends on conformational constraints
J. Biol. Chem., 292, 2017
|
|
