3LP9
 
 | | Crystal structure of LS24, A Seed Albumin from Lathyrus sativus | | Descriptor: | CALCIUM ION, CHLORIDE ION, LS-24, ... | | Authors: | Gaur, V, Qureshi, I.A, Singh, A, Chanana, V, Salunke, D.M. | | Deposit date: | 2010-02-05 | | Release date: | 2010-02-23 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal structure and functional insights of hemopexin fold protein from grass pea
Plant Physiol., 152, 2010
|
|
3OYO
 
 | | Crystal structure of hemopexin fold protein CP4 from cow pea | | Descriptor: | CALCIUM ION, CHLORIDE ION, SODIUM ION, ... | | Authors: | Gaur, V, Chanana, V, Salunke, D.M. | | Deposit date: | 2010-09-23 | | Release date: | 2011-02-09 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | The structure of a haemopexin-fold protein from cow pea (Vigna unguiculata) suggests functional diversity of haemopexins in plants
Acta Crystallogr.,Sect.F, 67, 2011
|
|
3EHK
 
 | |
4QWD
 
 | |
4XLG
 
 | | C. glabrata Slx1 in complex with Slx4CCD. | | Descriptor: | CHLORIDE ION, Structure-specific endonuclease subunit SLX1, Structure-specific endonuclease subunit SLX4, ... | | Authors: | Gaur, V, Wyatt, H.D.M, Komorowska, W, Szczepanowski, R.H, de Sanctis, D, Gorecka, K.M, West, S.C, Nowotny, M. | | Deposit date: | 2015-01-13 | | Release date: | 2015-03-25 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.78 Å) | | Cite: | Structural and Mechanistic Analysis of the Slx1-Slx4 Endonuclease.
Cell Rep, 10, 2015
|
|
4XM5
 
 | | C. glabrata Slx1. | | Descriptor: | CHLORIDE ION, Structure-specific endonuclease subunit SLX1, ZINC ION | | Authors: | Gaur, V, Wyatt, H.D.M, Komorowska, W, Szczepanowski, R.H, de Sanctis, D, Gorecka, K.M, West, S.C, Nowotny, M. | | Deposit date: | 2015-01-14 | | Release date: | 2015-03-25 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.34 Å) | | Cite: | Structural and Mechanistic Analysis of the Slx1-Slx4 Endonuclease.
Cell Rep, 10, 2015
|
|
6SEH
 
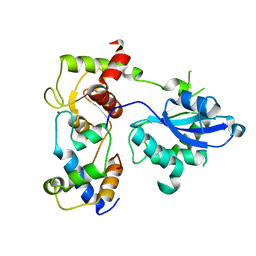 | | Recognition and processing of branched DNA substrates by Slx1-Slx4 nuclease | | Descriptor: | Structure-specific endonuclease subunit SLX1, Structure-specific endonuclease subunit SLX4, ZINC ION | | Authors: | Gaur, V, Zajko, W, Nirwal, S, Szlachcic, A, Gapinska, M, Nowotny, M. | | Deposit date: | 2019-07-30 | | Release date: | 2019-09-25 | | Last modified: | 2024-05-15 | | Method: | X-RAY DIFFRACTION (3.15 Å) | | Cite: | Recognition and processing of branched DNA substrates by Slx1-Slx4 nuclease.
Nucleic Acids Res., 47, 2019
|
|
6SEI
 
 | | Recognition and processing of branched DNA substrates by Slx1-Slx4 nuclease | | Descriptor: | CALCIUM ION, DNA (32-MER), Structure-specific endonuclease subunit SLX1, ... | | Authors: | Gaur, V, Zajko, W, Nirwal, S, Szlachcic, A, Gapinska, M, Nowotny, M. | | Deposit date: | 2019-07-30 | | Release date: | 2019-09-25 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.69 Å) | | Cite: | Recognition and processing of branched DNA substrates by Slx1-Slx4 nuclease.
Nucleic Acids Res., 47, 2019
|
|
4QW8
 
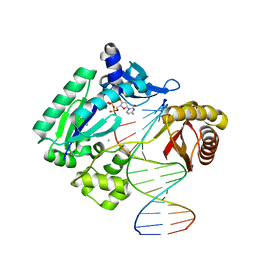 | |
4QWA
 
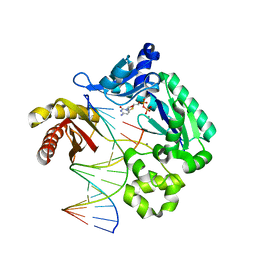 | |
4QW9
 
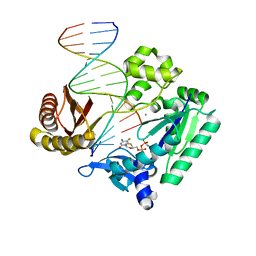 | |
4QWE
 
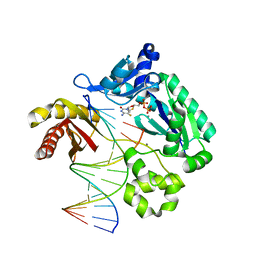 | |
7WME
 
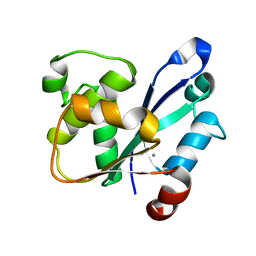 | | Crystal Structure of the catalytic domain of At-HIGLE | | Descriptor: | CALCIUM ION, Structure-specific endonuclease subunit SLX1 homolog | | Authors: | Verma, P, Kumari, P, Negi, S, Yadav, G, Gaur, V. | | Deposit date: | 2022-01-14 | | Release date: | 2022-04-13 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Holliday junction resolution by At-HIGLE: an SLX1 lineage endonuclease from Arabidopsis thaliana with a novel in-built regulatory mechanism.
Nucleic Acids Res., 50, 2022
|
|
8JSB
 
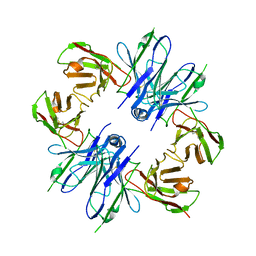 | |
8H73
 
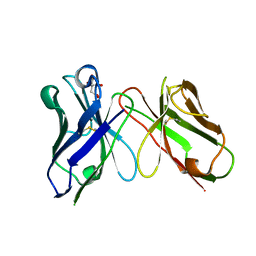 | | Crystal structure of antibody scFv against M2e Influenza peptide | | Descriptor: | CITRATE ANION, Single Chain Variable Fragment | | Authors: | Kumar, U, Madni, Z.K, Gaur, V, Salunke, D.M. | | Deposit date: | 2022-10-18 | | Release date: | 2023-08-02 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.91 Å) | | Cite: | A structure and knowledge-based combinatorial approach to engineering universal scFv antibodies against influenza M2 protein.
J.Biomed.Sci., 30, 2023
|
|
8H3C
 
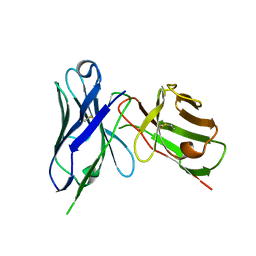 | | Crystal structure of M2e Influenza peptide in complex with antibody scFv | | Descriptor: | Matrix protein 2, Single Chain Variable Fragment | | Authors: | Kumar, U, Madni, Z.K, Gaur, V, Salunke, D.M. | | Deposit date: | 2022-10-08 | | Release date: | 2023-08-02 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (3.43 Å) | | Cite: | A structure and knowledge-based combinatorial approach to engineering universal scFv antibodies against influenza M2 protein.
J.Biomed.Sci., 30, 2023
|
|
8H3B
 
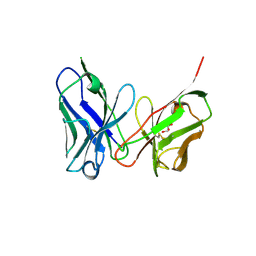 | | Crystal structure of antibody scFv against M2e Influenza peptide | | Descriptor: | GLYCEROL, Single Chain Variable Fragment | | Authors: | Kumar, U, Madni, Z.K, Gaur, V, Salunke, D.M. | | Deposit date: | 2022-10-08 | | Release date: | 2023-08-02 | | Method: | X-RAY DIFFRACTION (2.75 Å) | | Cite: | A structure and knowledge-based combinatorial approach to engineering universal scFv antibodies against influenza M2 protein.
J.Biomed.Sci., 30, 2023
|
|
7YUE
 
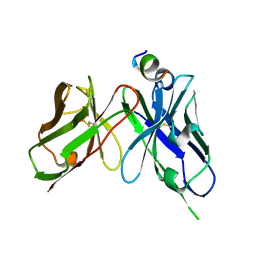 | | Epitope-directed anti-SARS CoV 2 scFv engineered against the key spike protein region. | | Descriptor: | Single chain variable Fragment, Spike protein S2 | | Authors: | Kumar, U, Jaiswal, D, Gaur, V, Salunke, D.M. | | Deposit date: | 2022-08-17 | | Release date: | 2023-02-08 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Epitope-directed anti-SARS-CoV-2 scFv engineered against the key spike protein region could block membrane fusion.
Protein Sci., 32, 2023
|
|
4H0I
 
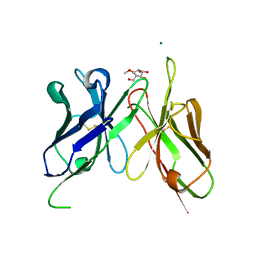 | | Crystal Structure of Scfv-2D10 in Complex with Methyl Alpha-D-Mannopyranoside | | Descriptor: | 2D10 scFv, MAGNESIUM ION, methyl alpha-D-mannopyranoside | | Authors: | Tapryal, S, Gaur, V, Kaur, K.J, Salunke, D.M. | | Deposit date: | 2012-09-08 | | Release date: | 2013-06-19 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structural evaluation of a mimicry-recognizing paratope: plasticity in antigen-antibody interactions manifests in molecular mimicry.
J.Immunol., 191, 2013
|
|
4H0G
 
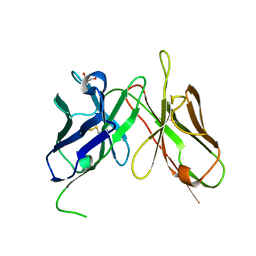 | | Crystal structure of mimicry-recognizing native 2D10 scFv | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, 2D10 scFv | | Authors: | Tapryal, S, Gaur, V, Kaur, K.J, Salunke, D.M. | | Deposit date: | 2012-09-08 | | Release date: | 2013-06-19 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structural evaluation of a mimicry-recognizing paratope: plasticity in antigen-antibody interactions manifests in molecular mimicry.
J.Immunol., 191, 2013
|
|
4H0H
 
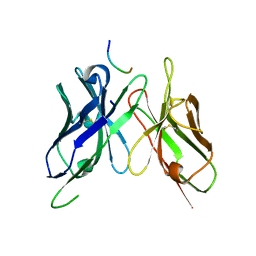 | | Crystal structure of mimicry-recognizing 2D10 scFv with peptide | | Descriptor: | 2D10 scFv, peptide | | Authors: | Tapryal, S, Gaur, V, Kaur, K.J, Salunke, D.M. | | Deposit date: | 2012-09-08 | | Release date: | 2013-06-19 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural evaluation of a mimicry-recognizing paratope: plasticity in antigen-antibody interactions manifests in molecular mimicry.
J.Immunol., 191, 2013
|
|
4QWB
 
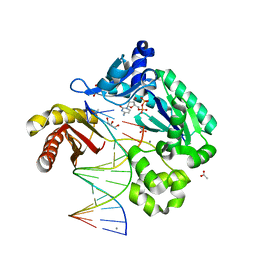 | |
4QWC
 
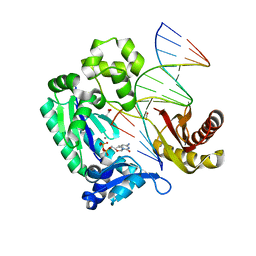 | |
5I4F
 
 | | scFv 2D10 complexed with alpha 1,6 mannobiose | | Descriptor: | alpha-D-mannopyranose-(1-6)-alpha-D-mannopyranose, scFv 2D10 | | Authors: | Vashisht, S, Kumar, A, Kaur, K.J, Salunke, D.M. | | Deposit date: | 2016-02-12 | | Release date: | 2016-12-21 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.549 Å) | | Cite: | Antibodies Can Exploit Molecular Crowding to Bind New Antigens at Noncanonical Paratope Positions
CHEMISTRYSELECT, 1, 2016
|
|
