6YPT
 
 | |
1ZXZ
 
 | | X-ray structure of peptide deformylase from Arabidopsis thaliana (AtPDF1A); crystals grown in PEG-5000 MME as precipitant | | Descriptor: | Peptide deformylase, mitochondrial, ZINC ION | | Authors: | Fieulaine, S, Juillan-Binard, C, Serero, A, Dardel, F, Giglione, C, Meinnel, T, Ferrer, J.-L. | | Deposit date: | 2005-06-09 | | Release date: | 2005-09-27 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | The crystal structure of mitochondrial (Type 1A) peptide deformylase provides clear guidelines for the design of inhibitors specific for the bacterial forms
J.Biol.Chem., 280, 2005
|
|
1ZY0
 
 | | X-ray structure of peptide deformylase from Arabidopsis thaliana (AtPDF1A); crystals grown in PEG-6000 | | Descriptor: | Peptide deformylase, mitochondrial, ZINC ION | | Authors: | Fieulaine, S, Juillan-Binard, C, Serero, A, Dardel, F, Giglione, C, Meinnel, T, Ferrer, J.-L. | | Deposit date: | 2005-06-09 | | Release date: | 2005-09-27 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | The crystal structure of mitochondrial (Type 1A) peptide deformylase provides clear guidelines for the design of inhibitors specific for the bacterial forms
J.Biol.Chem., 280, 2005
|
|
5JF4
 
 | | Crystal structure of type 2 PDF from Streptococcus agalactiae in complex with inhibitor AT019 | | Descriptor: | (3R)-3-{3-[(1-benzofuran-3-yl)methyl]-1,2,4-oxadiazol-5-yl}-4-cyclopentyl-N-hydroxybutanamide, ACETATE ION, IMIDAZOLE, ... | | Authors: | Fieulaine, S, Giglione, C, Meinnel, T. | | Deposit date: | 2016-04-19 | | Release date: | 2016-11-30 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | A unique peptide deformylase platform to rationally design and challenge novel active compounds.
Sci Rep, 6, 2016
|
|
5JF5
 
 | | Crystal structure of type 2 PDF from Streptococcus agalactiae in complex with inhibitor AT020 | | Descriptor: | (3R)-3-{3-[(2H-1,3-benzodioxol-5-yl)methyl]-1,2,4-oxadiazol-5-yl}-4-cyclopentyl-N-hydroxybutanamide, ACETATE ION, IMIDAZOLE, ... | | Authors: | Fieulaine, S, Giglione, C, Meinnel, T. | | Deposit date: | 2016-04-19 | | Release date: | 2016-11-30 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | A unique peptide deformylase platform to rationally design and challenge novel active compounds.
Sci Rep, 6, 2016
|
|
5JF0
 
 | | Crystal structure of type 2 PDF from Streptococcus agalactiae in complex with tripeptide Met-Ala-Arg | | Descriptor: | ACETATE ION, MET-ALA-ARG, NICKEL (II) ION, ... | | Authors: | Fieulaine, S, Giglione, C, Meinnel, T. | | Deposit date: | 2016-04-19 | | Release date: | 2016-11-30 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | A unique peptide deformylase platform to rationally design and challenge novel active compounds.
Sci Rep, 6, 2016
|
|
5JF2
 
 | | Crystal structure of type 2 PDF from Streptococcus agalactiae in complex with inhibitor AT002 | | Descriptor: | (3R)-3-{3-[(4-fluorophenyl)methyl]-1,2,4-oxadiazol-5-yl}-N-hydroxyheptanamide, ACETATE ION, IMIDAZOLE, ... | | Authors: | Fieulaine, S, Giglione, C, Meinnel, T, Hamiche, K. | | Deposit date: | 2016-04-19 | | Release date: | 2016-11-30 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | A unique peptide deformylase platform to rationally design and challenge novel active compounds.
Sci Rep, 6, 2016
|
|
1ZY1
 
 | | X-ray structure of peptide deformylase from Arabidopsis thaliana (AtPDF1A) in complex with Met-Ala-Ser | | Descriptor: | Peptide deformylase, mitochondrial, ZINC ION, ... | | Authors: | Fieulaine, S, Juillan-Binard, C, Serero, A, Dardel, F, Giglione, C, Meinnel, T, Ferrer, J.-L. | | Deposit date: | 2005-06-09 | | Release date: | 2005-09-27 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | The crystal structure of mitochondrial (Type 1A) peptide deformylase provides clear guidelines for the design of inhibitors specific for the bacterial forms
J.Biol.Chem., 280, 2005
|
|
1JB1
 
 | | Lactobacillus casei HprK/P Bound to Phosphate | | Descriptor: | HPRK PROTEIN, PHOSPHATE ION | | Authors: | Fieulaine, S, Morera, S, Poncet, S, Monedero, V, Gueguen-Chaignon, V, Galinier, A, Janin, J, Deutscher, J, Nessler, S. | | Deposit date: | 2001-06-01 | | Release date: | 2001-08-08 | | Last modified: | 2017-10-04 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | X-ray structure of HPr kinase: a bacterial protein kinase with a P-loop nucleotide-binding domain.
EMBO J., 20, 2001
|
|
1KKL
 
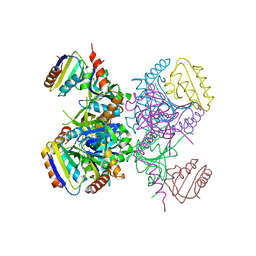 | | L.casei HprK/P in complex with B.subtilis HPr | | Descriptor: | CALCIUM ION, HprK protein, PHOSPHOCARRIER PROTEIN HPR | | Authors: | Fieulaine, S, Morera, S, Poncet, S, Galinier, A, Janin, J, Deutscher, J, Nessler, S. | | Deposit date: | 2001-12-10 | | Release date: | 2002-08-28 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | X-ray structure of a bifunctional protein kinase in complex with its protein substrate HPr.
Proc.Natl.Acad.Sci.USA, 99, 2002
|
|
1KKM
 
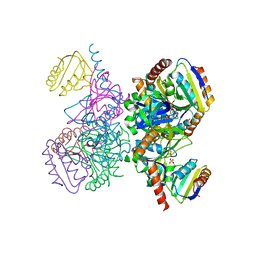 | | L.casei HprK/P in complex with B.subtilis P-Ser-HPr | | Descriptor: | CALCIUM ION, HprK protein, PHOSPHATE ION, ... | | Authors: | Fieulaine, S, Morera, S, Poncet, S, Galinier, A, Janin, J, Deutscher, J, Nessler, S. | | Deposit date: | 2001-12-10 | | Release date: | 2002-08-28 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | X-ray structure of a bifunctional protein kinase in complex with its protein substrate HPr.
Proc.Natl.Acad.Sci.USA, 99, 2002
|
|
4JE8
 
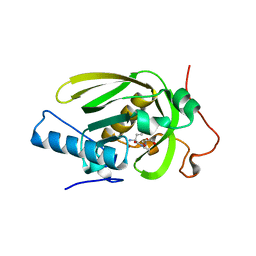 | | Crystal structure of a human-like mitochondrial peptide deformylase in complex with Met-Ala-Ser | | Descriptor: | Peptide deformylase 1A, chloroplastic/mitochondrial, ZINC ION, ... | | Authors: | Fieulaine, S, Meinnel, T, Giglione, C. | | Deposit date: | 2013-02-26 | | Release date: | 2014-02-26 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Understanding the highly efficient catalysis of prokaryotic peptide deformylases by shedding light on the determinants specifying the low activity of the human counterpart.
Acta Crystallogr.,Sect.D, 70, 2014
|
|
4JE7
 
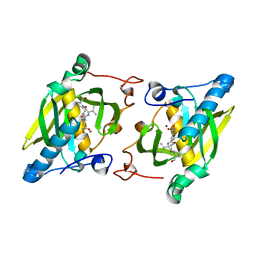 | | Crystal structure of a human-like mitochondrial peptide deformylase in complex with actinonin | | Descriptor: | ACTINONIN, Peptide deformylase 1A, chloroplastic/mitochondrial, ... | | Authors: | Fieulaine, S, Meinnel, T, Giglione, C. | | Deposit date: | 2013-02-26 | | Release date: | 2014-02-26 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Understanding the highly efficient catalysis of prokaryotic peptide deformylases by shedding light on the determinants specifying the low activity of the human counterpart.
Acta Crystallogr.,Sect.D, 70, 2014
|
|
4JE6
 
 | | Crystal structure of a human-like mitochondrial peptide deformylase | | Descriptor: | Peptide deformylase 1A, chloroplastic/mitochondrial, ZINC ION | | Authors: | Fieulaine, S, Meinnel, T, Giglione, C. | | Deposit date: | 2013-02-26 | | Release date: | 2014-02-26 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Understanding the highly efficient catalysis of prokaryotic peptide deformylases by shedding light on the determinants specifying the low activity of the human counterpart.
Acta Crystallogr.,Sect.D, 70, 2014
|
|
2D2V
 
 | |
2B1R
 
 | |
2B1Q
 
 | |
3M6O
 
 | |
3M6R
 
 | |
3M6P
 
 | |
3M6Q
 
 | |
3O3J
 
 | |
3PN2
 
 | |
3PN4
 
 | | Crystal structure of Arabidopsis thaliana petide deformylase 1B (AtPDF1B) in complex with actinonin (crystallized in PEG-550-MME) | | Descriptor: | ACTINONIN, Peptide deformylase 1B, chloroplastic, ... | | Authors: | Fieulaine, S, Meinnel, T, Giglione, C. | | Deposit date: | 2010-11-18 | | Release date: | 2011-06-08 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Trapping conformational States along ligand-binding dynamics of Peptide deformylase: the impact of induced fit on enzyme catalysis.
Plos Biol., 9, 2011
|
|
3PN5
 
 | |
