4V5B
 
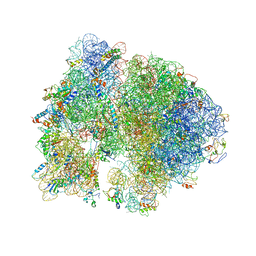 | | Structure of PDF binding helix in complex with the ribosome. | | Descriptor: | 16S RIBOSOMAL RNA, 23S RIBOSOMAL RNA, 30S RIBOSOMAL PROTEIN S10, ... | | Authors: | Bingel-Erlenmeyer, R, Kohler, R, Kramer, G, Sandikci, A, Antolic, S, Maier, T, Schaffitzel, C, Wiedmann, B, Bukau, B, Ban, N. | | Deposit date: | 2007-11-22 | | Release date: | 2014-07-09 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (3.74 Å) | | Cite: | A Peptide Deformylase-Ribosome Complex Reveals Mechanism of Nascent Chain Processing.
Nature, 452, 2008
|
|
2VRH
 
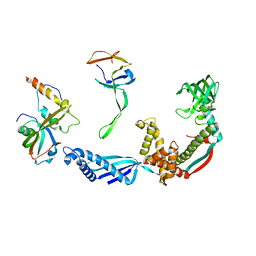 | | Structure of the E. coli trigger factor bound to a translating ribosome | | Descriptor: | 50S RIBOSOMAL PROTEIN L23, 50S RIBOSOMAL PROTEIN L24, 50S RIBOSOMAL PROTEIN L29, ... | | Authors: | Merz, F, Boehringer, D, Schaffitzel, C, Preissler, S, Hoffmann, A, Maier, T, Rutkowska, A, Lozza, J, Ban, N, Bukau, B, Deuerling, E. | | Deposit date: | 2008-04-07 | | Release date: | 2008-06-17 | | Last modified: | 2024-05-08 | | Method: | ELECTRON MICROSCOPY (19 Å) | | Cite: | Molecular Mechanism and Structure of Trigger Factor Bound to the Translating Ribosome.
Embo J., 27, 2008
|
|
1LZW
 
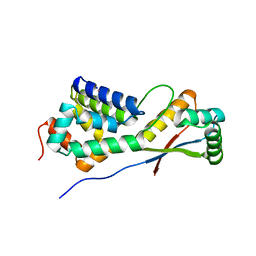 | | Structural basis of ClpS-mediated switch in ClpA substrate recognition | | Descriptor: | ATP-dependent clp protease ATP-binding subunit ClpA, PLATINUM (II) ION, Protein yljA | | Authors: | Zeth, K, Ravelli, R.B, Paal, K, Cusack, S, Bukau, B, Dougan, D.A. | | Deposit date: | 2002-06-11 | | Release date: | 2002-11-27 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structural analysis of the adaptor protein ClpS in complex with the N-terminal domain of ClpA
Nat.Struct.Biol., 9, 2002
|
|
1W2B
 
 | | Trigger Factor ribosome binding domain in complex with 50S | | Descriptor: | 23S RRNA, 50S RIBOSOMAL PROTEIN L10E, 50S RIBOSOMAL PROTEIN L13P, ... | | Authors: | Ferbitz, L, Maier, T, Patzelt, H, Bukau, B, Deuerling, E, Ban, N. | | Deposit date: | 2004-07-01 | | Release date: | 2004-09-02 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (3.5 Å) | | Cite: | Trigger Factor in Complex with the Ribosome Forms a Molecular Cradle for Nascent Proteins
Nature, 431, 2004
|
|
1W26
 
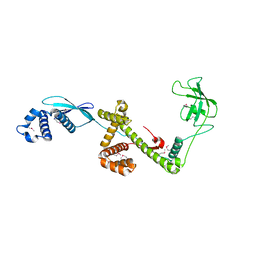 | | Trigger Factor in Complex with the Ribosome forms a Molecular Cradle for Nascent Proteins | | Descriptor: | TRIGGER FACTOR | | Authors: | Ferbitz, L, Maier, T, Patzelt, H, Bukau, B, Deuerling, E, Ban, N. | | Deposit date: | 2004-06-28 | | Release date: | 2004-09-02 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Trigger Factor in Complex with the Ribosome Forms a Molecular Cradle for Nascent Proteins
Nature, 431, 2004
|
|
6EM9
 
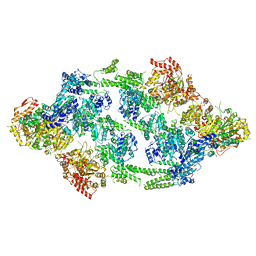 | | S.aureus ClpC resting state, asymmetric map | | Descriptor: | ATP-dependent Clp protease ATP-binding subunit ClpC | | Authors: | Carroni, M, Mogk, A, Bukau, B, Franke, K. | | Deposit date: | 2017-10-01 | | Release date: | 2017-12-27 | | Last modified: | 2024-05-15 | | Method: | ELECTRON MICROSCOPY (8.4 Å) | | Cite: | Regulatory coiled-coil domains promote head-to-head assemblies of AAA+ chaperones essential for tunable activity control.
Elife, 6, 2017
|
|
6EMW
 
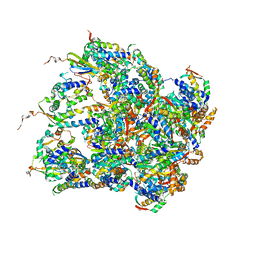 | | Structure of S.aureus ClpC in complex with MecA | | Descriptor: | ATP-dependent Clp protease ATP-binding subunit, ATP-dependent Clp protease ATP-binding subunit ClpC, Adapter protein MecA, ... | | Authors: | Carroni, M, Mogk, A, Bukau, B, Franke, K. | | Deposit date: | 2017-10-03 | | Release date: | 2017-12-27 | | Last modified: | 2024-05-15 | | Method: | ELECTRON MICROSCOPY (11 Å) | | Cite: | Regulatory coiled-coil domains promote head-to-head assemblies of AAA+ chaperones essential for tunable activity control.
Elife, 6, 2017
|
|
5OFO
 
 | | Cryo EM structure of the E. coli disaggregase ClpB (BAP form, DWB mutant), in the ATPgammaS state, bound to the model substrate casein | | Descriptor: | Chaperone protein ClpB,ATP-dependent Clp protease ATP-binding subunit ClpA,Chaperone protein ClpB, PHOSPHOTHIOPHOSPHORIC ACID-ADENYLATE ESTER | | Authors: | Deville, C, Carroni, M, Franke, K.B, Topf, M, Bukau, B, Mogk, A, Saibil, H.R. | | Deposit date: | 2017-07-11 | | Release date: | 2017-08-16 | | Last modified: | 2024-05-08 | | Method: | ELECTRON MICROSCOPY (4.6 Å) | | Cite: | Structural pathway of regulated substrate transfer and threading through an Hsp100 disaggregase.
Sci Adv, 3, 2017
|
|
5OG1
 
 | | Cryo EM structure of the E. coli disaggregase ClpB (BAP form, DWB mutant), in the ATPgammaS state | | Descriptor: | Chaperone protein ClpB,ATP-dependent Clp protease ATP-binding subunit ClpA,Chaperone protein ClpB, PHOSPHOTHIOPHOSPHORIC ACID-ADENYLATE ESTER | | Authors: | Deville, C, Carroni, M, Franke, K.B, Topf, M, Bukau, B, Mogk, A, Saibil, H.R. | | Deposit date: | 2017-07-11 | | Release date: | 2017-08-16 | | Last modified: | 2024-05-08 | | Method: | ELECTRON MICROSCOPY (4.5 Å) | | Cite: | Structural pathway of regulated substrate transfer and threading through an Hsp100 disaggregase.
Sci Adv, 3, 2017
|
|
1MG9
 
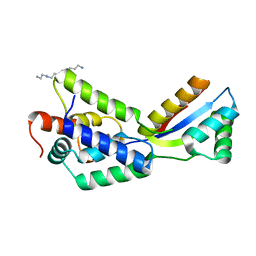 | | The structural basis of ClpS-mediated switch in ClpA substrate recognition | | Descriptor: | ATP dependent clp protease ATP-binding subunit clpA, SPERMINE (FULLY PROTONATED FORM), protein yljA | | Authors: | Zeth, K, Ravelli, R.B, Paal, K, Cusack, S, Bukau, B, Dougan, D.A. | | Deposit date: | 2002-08-15 | | Release date: | 2002-11-27 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structural analysis of the adaptor protein ClpS in complex with the N-terminal domain of ClpA
Nat.Struct.Biol., 9, 2002
|
|
4CIU
 
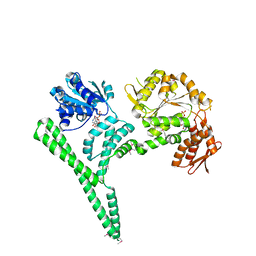 | | Crystal structure of E. coli ClpB | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, CHAPERONE PROTEIN CLPB | | Authors: | Kopp, J, Sinning, I, Bukau, B, Kummer, E, Mogk, A. | | Deposit date: | 2013-12-16 | | Release date: | 2014-05-14 | | Last modified: | 2019-05-22 | | Method: | X-RAY DIFFRACTION (3.5 Å) | | Cite: | Head-to-Tail Interactions of the Coiled-Coil Domains Regulate Clpb Cooperation with Hsp70 in Protein Disaggregation
Elife, 3, 2014
|
|
4D2U
 
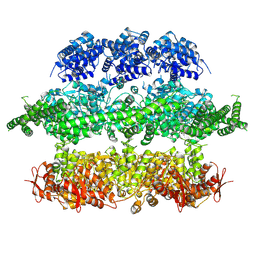 | | Negative-stain electron microscopy of E. coli ClpB (BAP form bound to ClpP) | | Descriptor: | CHAPERONE PROTEIN CLPB | | Authors: | Carroni, M, Kummer, E, Oguchi, Y, Clare, D.K, Wendler, P, Sinning, I, Kopp, J, Mogk, A, Bukau, B, Saibil, H.R. | | Deposit date: | 2014-05-13 | | Release date: | 2014-06-04 | | Last modified: | 2017-08-23 | | Method: | ELECTRON MICROSCOPY (17 Å) | | Cite: | Head-to-Tail Interactions of the Coiled-Coil Domains Regulate Clpb Activity and Cooperation with Hsp70 in Protein Disaggregation.
Elife, 3, 2014
|
|
4D2Q
 
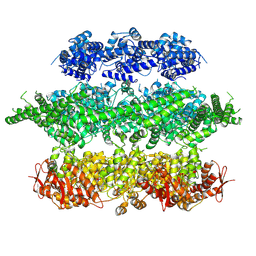 | | Negative-stain electron microscopy of E. coli ClpB mutant E432A (BAP form bound to ClpP) | | Descriptor: | CLPB | | Authors: | Carroni, M, Kummer, E, Oguchi, Y, Clare, D.K, Wendler, P, Sinning, I, Kopp, J, Mogk, A, Bukau, B, Saibil, H.R. | | Deposit date: | 2014-05-12 | | Release date: | 2014-06-04 | | Last modified: | 2017-08-23 | | Method: | ELECTRON MICROSCOPY (18 Å) | | Cite: | Head-to-Tail Interactions of the Coiled-Coil Domains Regulate Clpb Activity and Cooperation with Hsp70 in Protein Disaggregation.
Elife, 3, 2014
|
|
4D2X
 
 | | Negative-stain electron microscopy of E. coli ClpB of Y503D hyperactive mutant (BAP form bound to ClpP) | | Descriptor: | CHAPERONE PROTEIN CLPB | | Authors: | Carroni, M, Kummer, E, Oguchi, Y, Clare, D.K, Wendler, P, Sinning, I, Kopp, J, Mogk, A, Bukau, B, Saibil, H.R. | | Deposit date: | 2014-05-13 | | Release date: | 2014-06-04 | | Last modified: | 2024-10-16 | | Method: | ELECTRON MICROSCOPY (20 Å) | | Cite: | Head-to-Tail Interactions of the Coiled-Coil Domains Regulate Clpb Activity and Cooperation with Hsp70 in Protein Disaggregation.
Elife, 3, 2014
|
|
6Z5N
 
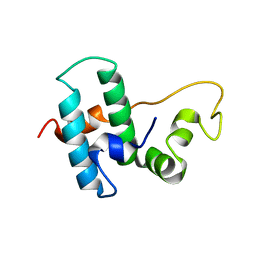 | | DnaJB1 JD-GF | | Descriptor: | DnaJ homolog subfamily B member 1 | | Authors: | Avraham-Abayev, M, London, N, Rosenzweig, R. | | Deposit date: | 2020-05-27 | | Release date: | 2020-11-18 | | Last modified: | 2024-06-19 | | Method: | SOLUTION NMR | | Cite: | HSP40 proteins use class-specific regulation to drive HSP70 functional diversity.
Nature, 587, 2020
|
|
6GOX
 
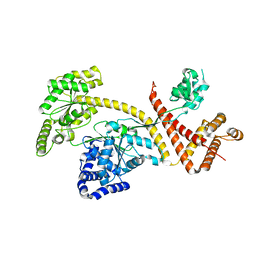 | | SecA | | Descriptor: | Protein translocase subunit SecA | | Authors: | White, S.A, Huber, D. | | Deposit date: | 2018-06-04 | | Release date: | 2019-06-19 | | Last modified: | 2024-05-15 | | Method: | X-RAY DIFFRACTION (3.5 Å) | | Cite: | The C-terminal tail of the bacterial translocation ATPase SecA modulates its activity.
Elife, 8, 2019
|
|
2BZT
 
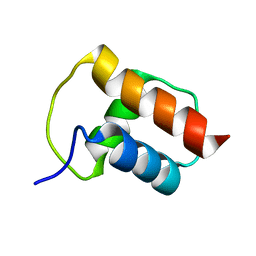 | | NMR structure of the bacterial protein YFHJ from E. coli | | Descriptor: | PROTEIN ISCX | | Authors: | Pastore, C, Kelly, G, Adinolfi, S, Mc Cormick, J.E, Pastore, A. | | Deposit date: | 2005-08-22 | | Release date: | 2006-12-06 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | YfhJ, a molecular adaptor in iron-sulfur cluster formation or a frataxin-like protein?
Structure, 14, 2006
|
|
4IO8
 
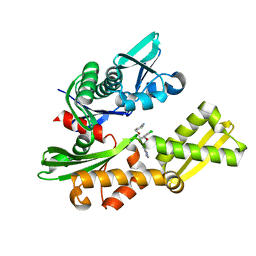 | | Crystal structure of human HSP70 complexed with 4-{(2R,3S,4R)-5-[(R)-6-Amino-8-(3,4-dichloro-benzylamino)-purin-9-yl]-3,4-dihydroxy-tetrahydro-furan-2-ylmethoxymethyl}-benzonitrile | | Descriptor: | 4-[[(2R,3S,4R,5R)-5-[6-amino-8-[(3,4-dichlorophenyl)methylamino]purin-9-yl]-3,4-dihydroxy-oxolan-2-yl]methoxymethyl]benzonitrile, Heat shock 70kDa protein 1A variant | | Authors: | Musil, D, Scholz, S. | | Deposit date: | 2013-01-07 | | Release date: | 2013-12-18 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.58 Å) | | Cite: | Functional analysis of hsp70 inhibitors.
Plos One, 8, 2013
|
|
6EM8
 
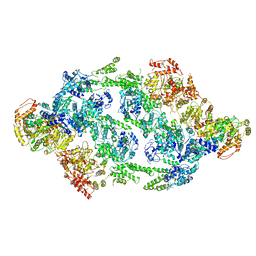 | | S.aureus ClpC resting state, C2 symmetrised | | Descriptor: | ATP-dependent Clp protease ATP-binding subunit ClpC | | Authors: | Carroni, M, Mogk, A. | | Deposit date: | 2017-10-01 | | Release date: | 2017-12-27 | | Last modified: | 2024-05-15 | | Method: | ELECTRON MICROSCOPY (8.4 Å) | | Cite: | Regulatory coiled-coil domains promote head-to-head assemblies of AAA+ chaperones essential for tunable activity control.
Elife, 6, 2017
|
|
6QS8
 
 | |
6QS4
 
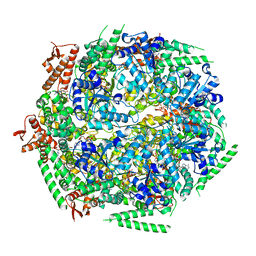 | |
6QS7
 
 | |
7QV3
 
 | |
7QV1
 
 | | Bacillus subtilis collided disome (Leading 70S) | | Descriptor: | 16S ribosomal RNA, 23S ribosomal RNA, 30S ribosomal protein S10, ... | | Authors: | Filbeck, S, Pfeffer, S. | | Deposit date: | 2022-01-19 | | Release date: | 2022-03-09 | | Last modified: | 2022-03-30 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | Bacterial ribosome collision sensing by a MutS DNA repair ATPase paralogue.
Nature, 603, 2022
|
|
7QV2
 
 | | Bacillus subtilis collided disome (Collided 70S) | | Descriptor: | 16S ribosomal RNA, 23S ribosomal RNA, 30S ribosomal protein S10, ... | | Authors: | Filbeck, S, Pfeffer, S. | | Deposit date: | 2022-01-19 | | Release date: | 2022-03-09 | | Last modified: | 2024-07-17 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | Bacterial ribosome collision sensing by a MutS DNA repair ATPase paralogue.
Nature, 603, 2022
|
|
