6E18
 
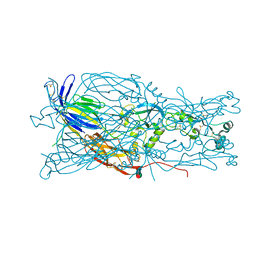 | | Crystal structure of Chlamydomonas reinhardtii HAP2 ectodomain provides structural insights of functional loops in green algae. | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, GLYCEROL, ... | | Authors: | Baquero, E, Legrand, P, Rey, F.A. | | Deposit date: | 2018-07-09 | | Release date: | 2018-11-07 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Species-Specific Functional Regions of the Green Alga Gamete Fusion Protein HAP2 Revealed by Structural Studies.
Structure, 27, 2019
|
|
5MDM
 
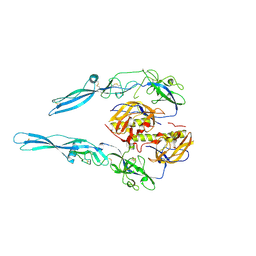 | | Structural intermediates in the fusion associated transition of vesiculovirus glycoprotein | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Baquero, E, Albertini, A.A, Gaudin, Y, Bressanelli, S. | | Deposit date: | 2016-11-11 | | Release date: | 2016-12-14 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.998 Å) | | Cite: | Structural intermediates in the fusion-associated transition of vesiculovirus glycoprotein.
EMBO J., 36, 2017
|
|
4D6W
 
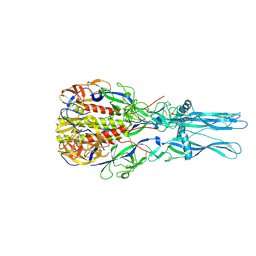 | | Crystal Structure of the low pH conformation of Chandipura Virus glycoprotein G ectodomain | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, GLYCOPROTEIN G, ... | | Authors: | Baquero, E, Albertini, A, Raux, H, Bressanelli, S, Gaudin, Y. | | Deposit date: | 2014-11-18 | | Release date: | 2015-03-04 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (3.6 Å) | | Cite: | Structure of the Low Ph Conformation of Chandipura Virus G Reveals Important Features in the Evolution of the Vesiculovirus Glycoprotein.
Plos Pathog., 11, 2015
|
|
7PKI
 
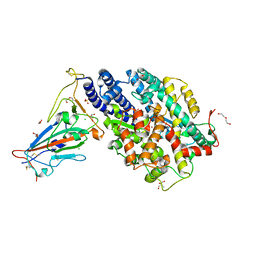 | |
8C8T
 
 | | cryo-EM structure of BG505 SOSIP.664 HIV-1 Env trimer in complex with bNAbs EPTC112 and 3BNC117 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Envelope glycoprotein gp160, ... | | Authors: | Baquero, E, Molinos-Albert, L, Mouquet, H. | | Deposit date: | 2023-01-21 | | Release date: | 2023-07-05 | | Last modified: | 2023-08-23 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Anti-V1/V3-glycan broadly HIV-1 neutralizing antibodies in a post-treatment controller.
Cell Host Microbe, 31, 2023
|
|
8S0N
 
 | |
8S0L
 
 | | Crystal structure of the TMPRSS2 zymogen in complex with the nanobody A07 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, Nanobody A07, ... | | Authors: | Duquerroy, S, Fernandez, I, Rey, F. | | Deposit date: | 2024-02-14 | | Release date: | 2024-06-26 | | Last modified: | 2024-09-04 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structural basis of TMPRSS2 zymogen activation and recognition by the HKU1 seasonal coronavirus.
Cell, 187, 2024
|
|
8S0M
 
 | | Crystal structure of the HKU1 receptor binding domain in complex with TMPRSS2 and the nanobody A01 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Nanobody A01, ... | | Authors: | Duquerroy, S, Fernandez, I, Rey, F. | | Deposit date: | 2024-02-14 | | Release date: | 2024-06-26 | | Last modified: | 2024-09-04 | | Method: | X-RAY DIFFRACTION (3.55 Å) | | Cite: | Structural basis of TMPRSS2 zymogen activation and recognition by the HKU1 seasonal coronavirus.
Cell, 187, 2024
|
|
6RX1
 
 | | Crystal structure of human syncytin 1 in post-fusion conformation | | Descriptor: | CHLORIDE ION, GLYCEROL, Syncytin-1 | | Authors: | Ruigrok, K, Backovic, M, Vaney, M.C, Rey, F.A. | | Deposit date: | 2019-06-07 | | Release date: | 2019-11-20 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | X-ray Structures of the Post-fusion 6-Helix Bundle of the Human Syncytins and their Functional Implications.
J.Mol.Biol., 431, 2019
|
|
6RX3
 
 | | Crystal structure of human syncytin 2 in post-fusion conformation | | Descriptor: | CHLORIDE ION, Syncytin-2 | | Authors: | Ruigrok, K, Backovic, M, Vaney, M.C, Rey, F.A. | | Deposit date: | 2019-06-07 | | Release date: | 2019-11-20 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | X-ray Structures of the Post-fusion 6-Helix Bundle of the Human Syncytins and their Functional Implications.
J.Mol.Biol., 431, 2019
|
|
