6SR7
 
 | | Structure of the U1A variant A1-98 Y31H/Q36R/K98W | | Descriptor: | SULFATE ION, U1 small nuclear ribonucleoprotein A | | Authors: | Rosenbach, H, Span, I. | | Deposit date: | 2019-09-05 | | Release date: | 2020-05-13 | | Last modified: | 2024-05-15 | | Method: | X-RAY DIFFRACTION (1.86 Å) | | Cite: | Expanding crystallization tools for nucleic acid complexes using U1A protein variants.
J.Struct.Biol., 210, 2020
|
|
6SQQ
 
 | |
6SQV
 
 | | Structure of the U1A variant A1-98 Y31H/Q36R/R70W | | Descriptor: | SULFATE ION, U1 small nuclear ribonucleoprotein A | | Authors: | Rosenbach, H, Span, I. | | Deposit date: | 2019-09-04 | | Release date: | 2020-05-13 | | Last modified: | 2024-05-15 | | Method: | X-RAY DIFFRACTION (2.45 Å) | | Cite: | Expanding crystallization tools for nucleic acid complexes using U1A protein variants.
J.Struct.Biol., 210, 2020
|
|
6SQT
 
 | |
6SQN
 
 | |
2JDF
 
 | | Human gamma-B crystallin | | Descriptor: | GAMMA CRYSTALLIN B | | Authors: | Ebersbach, H, Fiedler, E, Scheuermann, T, Fiedler, M, Stubbs, M.T, Reimann, C, Proetzel, G, Rudolph, R, Fiedler, U. | | Deposit date: | 2007-01-08 | | Release date: | 2007-07-24 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Affilin-Novel Binding Molecules Based on Human Gamma-B-Crystallin, an All Beta-Sheet Protein.
J.Mol.Biol., 372, 2007
|
|
2JDG
 
 | | Affilin based on HUMAN GAMMA-B CRYSTALLIN | | Descriptor: | GAMMA CRYSTALLIN B | | Authors: | Ebersbach, H, Fiedler, E, Scheuermann, T, Fiedler, M, Stubbs, M.T, Reimann, C, Proetzel, G, Rudolph, R, Fiedler, U. | | Deposit date: | 2007-01-08 | | Release date: | 2007-07-24 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Affilin-Novel Binding Molecules Based on Human Gamma-B-Crystallin, an All Beta-Sheet Protein.
J.Mol.Biol., 372, 2007
|
|
1ODO
 
 | | 1.85 A structure of CYP154A1 from Streptomyces coelicolor A3(2) | | Descriptor: | 4-PHENYL-1H-IMIDAZOLE, PROTOPORPHYRIN IX CONTAINING FE, PUTATIVE CYTOCHROME P450 154A1 | | Authors: | Podust, L.M, Kim, Y, Arase, M, Bach, H, Sherman, D.H, Lamb, D.C, Kelly, S.L, Waterman, M.R. | | Deposit date: | 2003-02-19 | | Release date: | 2004-01-02 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Comparison of the 1.85 A Structure of Cyp154A1 from Streptomyces Coelicolor A3(2) with the Closely Related Cyp154C1 and Cyps from Antibiotic Biosynthetic Pathways.
Protein Sci., 13, 2004
|
|
1GWI
 
 | | The 1.92 A structure of Streptomyces coelicolor A3(2) CYP154C1: A new monooxygenase that functionalizes macrolide ring systems | | Descriptor: | CYTOCHROME P450 154C1, PROTOPORPHYRIN IX CONTAINING FE, SULFATE ION | | Authors: | Podust, L.M, Kim, Y, Arase, M, Neely, B.A, Beck, B.J, Bach, H, Sherman, D.H, Lamb, D.C, Kelly, S.L, Waterman, M.R. | | Deposit date: | 2002-03-15 | | Release date: | 2003-01-29 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.92 Å) | | Cite: | The 1.92 A Structure of Streptomyces Coelicolor A3(2) Cyp154C1: A New Monooxygenase that Functionalizes Macrolide Ring Systems
J.Biol.Chem., 278, 2003
|
|
1FVG
 
 | | CRYSTAL STRUCTURE OF BOVINE PEPTIDE METHIONINE SULFOXIDE REDUCTASE | | Descriptor: | 2,3-DIHYDROXY-1,4-DITHIOBUTANE, PEPTIDE METHIONINE SULFOXIDE REDUCTASE | | Authors: | Lowther, W.T, Brot, N, Weissbach, H, Matthews, B.W. | | Deposit date: | 2000-09-19 | | Release date: | 2000-11-08 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Structure and mechanism of peptide methionine sulfoxide reductase, an "anti-oxidation" enzyme.
Biochemistry, 39, 2000
|
|
1FVA
 
 | | CRYSTAL STRUCTURE OF BOVINE METHIONINE SULFOXIDE REDUCTASE | | Descriptor: | PEPTIDE METHIONINE SULFOXIDE REDUCTASE | | Authors: | Lowther, W.T, Brot, N, Weissbach, H, Matthews, B.W. | | Deposit date: | 2000-09-19 | | Release date: | 2000-11-08 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structure and mechanism of peptide methionine sulfoxide reductase, an "anti-oxidation" enzyme.
Biochemistry, 39, 2000
|
|
6YM5
 
 | | Crystal structure of BAY-091 with PIP4K2A | | Descriptor: | (2~{R})-2-[[3-cyano-2-[4-(2-fluoranyl-3-methyl-phenyl)phenyl]-1,7-naphthyridin-4-yl]amino]butanoic acid, PHOSPHATE ION, Phosphatidylinositol 5-phosphate 4-kinase type-2 alpha | | Authors: | Holton, S.J, Wortmann, L, Braeuer, N, Irlbacher, H, Weiske, J, Lechner, C, Meier, R, Puetter, V, Christ, C, ter Laak, T, Lienau, P, Lesche, R, Nicke, B, Bauser, M, Haegebarth, A, von Nussbaum, F, Mumberg, D, Lemos, C. | | Deposit date: | 2020-04-07 | | Release date: | 2021-04-14 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Discovery and Characterization of the Potent and Highly Selective 1,7-Naphthyridine-Based Inhibitors BAY-091 and BAY-297 of the Kinase PIP4K2A.
J.Med.Chem., 64, 2021
|
|
6YM3
 
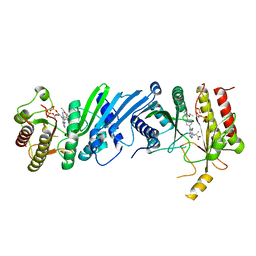 | | Crystal structure of Compound 1 with PIP4K2A | | Descriptor: | (2~{R})-2-[[3-cyano-2-[4-(2-ethoxyphenyl)phenyl]-5,8-dihydro-1,7-naphthyridin-4-yl]amino]propanoic acid, PHOSPHATE ION, Phosphatidylinositol 5-phosphate 4-kinase type-2 alpha | | Authors: | Holton, S.J, Wortmann, L, Braeuer, N, Irlbacher, H, Weiske, J, Lechner, C, Meier, R, Puetter, V, Christ, C, ter Laak, T, Lienau, P, Lesche, R, Nicke, B, Bauser, M, Haegebarth, A, von Nussbaum, F, Mumberg, D, Lemos, C. | | Deposit date: | 2020-04-07 | | Release date: | 2021-04-14 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Discovery and Characterization of the Potent and Highly Selective 1,7-Naphthyridine-Based Inhibitors BAY-091 and BAY-297 of the Kinase PIP4K2A.
J.Med.Chem., 64, 2021
|
|
6YM4
 
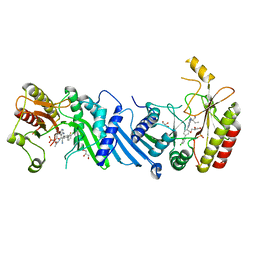 | | Crystal structure of BAY-297 with PIP4K2A | | Descriptor: | (2~{R})-2-[[2-[4-(3-chloranyl-2-fluoranyl-phenyl)phenyl]-3-cyano-1,7-naphthyridin-4-yl]amino]butanamide, GLYCEROL, Phosphatidylinositol 5-phosphate 4-kinase type-2 alpha | | Authors: | Holton, S.J, Wortmann, L, Braeuer, N, Irlbacher, H, Weiske, J, Lechner, C, Meier, R, Puetter, V, Christ, C, ter Laak, T, Lienau, P, Lesche, R, Nicke, B, Bauser, M, Haegebarth, A, von Nussbaum, F, Mumberg, D, Lemos, C. | | Deposit date: | 2020-04-07 | | Release date: | 2021-04-14 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Discovery and Characterization of the Potent and Highly Selective 1,7-Naphthyridine-Based Inhibitors BAY-091 and BAY-297 of the Kinase PIP4K2A.
J.Med.Chem., 64, 2021
|
|
1B4G
 
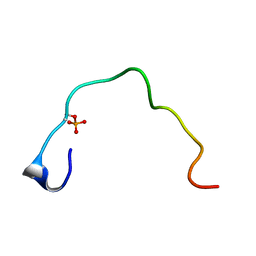 | | CONTROL OF K+ CHANNEL GATING BY PROTEIN PHOSPHORYLATION: STRUCTURAL SWITCHES OF THE INACTIVATION GATE, NMR, 22 STRUCTURES | | Descriptor: | POTASSIUM CHANNEL | | Authors: | Antz, C, Bauer, T, Kalbacher, H, Frank, R, Covarrubias, M, Kalbitzer, H.R, Ruppersberg, J.P, Baukrowitz, T, Fakler, B. | | Deposit date: | 1998-12-22 | | Release date: | 1999-04-27 | | Last modified: | 2024-10-09 | | Method: | SOLUTION NMR | | Cite: | Control of K+ channel gating by protein phosphorylation: structural switches of the inactivation gate.
Nat.Struct.Biol., 6, 1999
|
|
1L1D
 
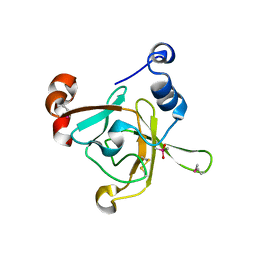 | | Crystal structure of the C-terminal methionine sulfoxide reductase domain (MsrB) of N. gonorrhoeae pilB | | Descriptor: | CACODYLATE ION, peptide methionine sulfoxide reductase | | Authors: | Lowther, W.T, Weissbach, H, Etienne, F, Brot, N, Matthews, B.W. | | Deposit date: | 2002-02-15 | | Release date: | 2002-05-01 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | The mirrored methionine sulfoxide reductases of Neisseria gonorrhoeae pilB.
Nat.Struct.Biol., 9, 2002
|
|
1B4I
 
 | | Control of K+ Channel Gating by protein phosphorylation: structural switches of the inactivation gate, NMR, 22 structures | | Descriptor: | POTASSIUM CHANNEL | | Authors: | Antz, C, Bauer, T, Kalbacher, H, Frank, R, Covarrubias, M, Kalbitzer, H.R, Ruppersberg, J.P, Baukrowitz, T, Fakler, B. | | Deposit date: | 1998-12-22 | | Release date: | 1999-04-27 | | Last modified: | 2024-10-23 | | Method: | SOLUTION NMR | | Cite: | Control of K+ channel gating by protein phosphorylation: structural switches of the inactivation gate.
Nat.Struct.Biol., 6, 1999
|
|
2XDP
 
 | | Crystal structure of the tudor domain of human JMJD2C | | Descriptor: | LYSINE-SPECIFIC DEMETHYLASE 4C, SULFATE ION | | Authors: | Yue, W.W, Gileadi, C, Krojer, T, Weisbach, H, Ugochukwu, E, Daniel, M, Phillips, C, Chaikuad, A, von Delft, F, Allerston, C, Arrowsmith, C, Weigelt, J, Edwards, A, Bountra, C, Oppermann, U. | | Deposit date: | 2010-05-06 | | Release date: | 2010-06-09 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.56 Å) | | Cite: | Crystal Structure of the Tudor Domain of Human Jmjd2C
To be Published
|
|
2H30
 
 | | Crystal structure of the N-terminal domain of PilB from Neisseria gonorrhoeae | | Descriptor: | Peptide methionine sulfoxide reductase msrA/msrB | | Authors: | Brot, N, Collet, J.F, Johnson, L.C, Jonsson, T.J, Weissbach, H, Lowther, W.T. | | Deposit date: | 2006-05-20 | | Release date: | 2006-08-22 | | Last modified: | 2021-10-20 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | The Thioredoxin Domain of Neisseria gonorrhoeae PilB Can Use Electrons from DsbD to Reduce Downstream Methionine Sulfoxide Reductases.
J.Biol.Chem., 281, 2006
|
|
2KB6
 
 | | Solution structure of onconase C87A/C104A | | Descriptor: | Protein P-30 | | Authors: | Weininger, U, Schulenburg, C, Arnold, U, Ulbrich-Hofmann, R, Balbach, J. | | Deposit date: | 2008-11-21 | | Release date: | 2009-11-24 | | Last modified: | 2024-10-30 | | Method: | SOLUTION NMR | | Cite: | Impact of the C-terminal disulfide bond on the folding and stability of onconase.
Chembiochem, 11, 2010
|
|
3FD7
 
 | | Crystal structure of Onconase C87A/C104A-ONC | | Descriptor: | 1,2-ETHANEDIOL, GLYCEROL, Protein P-30, ... | | Authors: | Neumann, P, Schulenburg, C, Arnold, U, Ulbrich-Hofmann, R, Stubbs, M.T. | | Deposit date: | 2008-11-25 | | Release date: | 2009-12-08 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.531 Å) | | Cite: | Impact of the C-terminal disulfide bond on the folding and stability of onconase.
Chembiochem, 11, 2010
|
|
7PDU
 
 | |
1W1F
 
 | | SH3 DOMAIN OF HUMAN LYN TYROSINE KINASE | | Descriptor: | TYROSINE-PROTEIN KINASE LYN | | Authors: | Bauer, F, Schweimer, K, Hoffmann, S, Roesch, P, Sticht, H. | | Deposit date: | 2004-06-17 | | Release date: | 2005-07-06 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Structural Characterization of Lyn-SH3 Domain in Complex with a Herpesviral Protein Reveals an Extended Recognition Motif that Enhances Binding Affinity.
Protein Sci., 14, 2005
|
|
1TBQ
 
 | |
1TBR
 
 | |
