4MQB
 
 | | Crystal structure of thymidylate kinase from Staphylococcus aureus in complex with 2-(N-morpholino)ethanesulfonic acid | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, TETRAETHYLENE GLYCOL, Thymidylate kinase | | Authors: | Filippova, E.V, Minasov, G, Shuvalova, L, Kiryukhina, O, Jedrzejczak, R, Babnigg, G, Rubin, E, Sacchettini, J, Joachimiak, A, Anderson, W.F, Midwest Center for Structural Genomics (MCSG), Structures of Mtb Proteins Conferring Susceptibility to Known Mtb Inhibitors (MTBI) | | Deposit date: | 2013-09-16 | | Release date: | 2013-10-23 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Crystal structure of thymidylate kinase from Staphylococcus aureus in complex with 2-(N-morpholino)ethanesulfonic acid
To be Published
|
|
5WHM
 
 | | Crystal Structure of IclR Family Transcriptional Regulator from Brucella abortus | | Descriptor: | 1,2-ETHANEDIOL, ACETIC ACID, CALCIUM ION, ... | | Authors: | Kim, Y, Wu, R, Tesar, C, Endres, M, Babnigg, G, Crosson, S, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2017-07-17 | | Release date: | 2017-08-23 | | Last modified: | 2019-11-27 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Molecular control of gene expression byBrucellaBaaR, an IclR-type transcriptional repressor.
J. Biol. Chem., 293, 2018
|
|
7L91
 
 | | Structure of Metallo Beta-Lactamase L1 in a Complex with Hydrolyzed Moxalactam Determined by Pink-Beam Serial Crystallography | | Descriptor: | (2R)-2-[(R)-carboxy{[(2R)-2-carboxy-2-(4-hydroxyphenyl)acetyl]amino}methoxymethyl]-5-{[(1-methyl-1H-tetrazol-5-yl)sulfanyl]methyl}-3,6-dihydro-2H-1,3-oxazine-4-carboxylic acid, Putative metallo-beta-lactamase l1 (Beta-lactamase type ii) (Ec 3.5.2.6) (Penicillinase), ZINC ION | | Authors: | Wilamowski, M, Kim, Y, Sherrell, D.A, Lavens, A, Henning, R, Maltseva, N, Endres, M, Babnigg, G, Vukica, S, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2021-01-01 | | Release date: | 2022-02-09 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Time-resolved beta-lactam cleavage by L1 metallo-beta-lactamase.
Nat Commun, 13, 2022
|
|
9D91
 
 | | Crystal structure of L-asparaginase from Streptococcus pneumoniae TIGR4 | | Descriptor: | 1,2-ETHANEDIOL, Asparaginase, SULFATE ION | | Authors: | Gade, P, Endres, M, Babnigg, G, Joachimiak, A, Center for Structural Biology of Infectious Diseases (CSBID) | | Deposit date: | 2024-08-20 | | Release date: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (1.62 Å) | | Cite: | Crystal structure of L-asparaginase from
Streptococcus pneumoniae TIGR4
To Be Published
|
|
5TF0
 
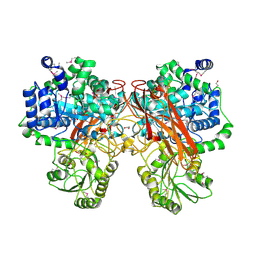 | | Crystal Structure of Glycosil Hydrolase Family 3 N-Terminal Domain Protein from Bacteroides intestinalis | | Descriptor: | 1,2-ETHANEDIOL, Glycosyl hydrolase family 3 N-terminal domain protein, MAGNESIUM ION | | Authors: | Kim, Y, Hatzos-Skintges, C, Endres, M, Babnigg, G, Joachimiak, A, MCSG, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2016-09-23 | | Release date: | 2016-10-05 | | Last modified: | 2019-12-25 | | Method: | X-RAY DIFFRACTION (2.2021 Å) | | Cite: | Crystal Structure of Glycosil Hydrolase Family 3 N-Terminal Domain Protein from Bacteroides intestinalis
To Be Published, 2016
|
|
7K3M
 
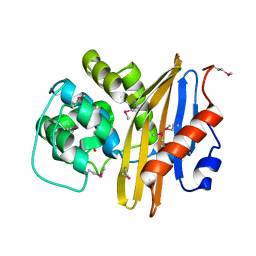 | | Crystal Structure of the Beta Lactamase Class D from Chitinophaga pinensis by Serial Crystallography | | Descriptor: | Beta-lactamase | | Authors: | Kim, Y, Sherrell, D.A, Johnson, J, Lavens, A, Maltseva, N, Endres, M, Babnigg, G, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2020-09-11 | | Release date: | 2020-09-23 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal Structure of the Beta Lactamase Class D from Chitinophaga pinensis by Serial Crystallography
To Be Published
|
|
7L52
 
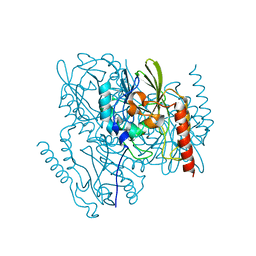 | | Crystal Structure of the Metallo Beta Lactamase L1 from Stenotrophomonas maltophilia Determined by Serial Crystallography | | Descriptor: | Putative metallo-beta-lactamase l1 (Beta-lactamase type ii) (Ec 3.5.2.6) (Penicillinase), ZINC ION | | Authors: | Wilamowski, M, Kim, Y, Sherrell, D.A, Lavens, A, Maltseva, N, Endres, M, Babnigg, G, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2020-12-21 | | Release date: | 2020-12-30 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Crystal Structure of the Metallo Beta Lactamase L1 from Stenotrophomonas maltophilia Determined by Serial Crystallography
To Be Published
|
|
3FRW
 
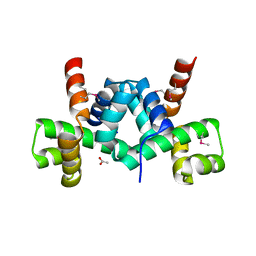 | | Crystal structure of putative TrpR protein from Ruminococcus obeum | | Descriptor: | ACETATE ION, Putative Trp repressor protein | | Authors: | Osipiuk, J, Keigher, L, Jedrzejczak, R, Babnigg, G, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2009-01-08 | | Release date: | 2009-01-20 | | Last modified: | 2020-01-22 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | X-ray crystal structure of putative TrpR protein from Ruminococcus obeum.
To be Published
|
|
3G1C
 
 | | The crystal structure of a TrpR like protein from Eubacterium eligens ATCC 27750 | | Descriptor: | MAGNESIUM ION, The TrpR like protein from Eubacterium eligens ATCC 27750 | | Authors: | Zhang, R, Hendricks, R, Freeman, L, Babnigg, G, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2009-01-29 | | Release date: | 2009-02-17 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | The crystal structure of a TrpR like protein from Eubacterium eligens ATCC 27750
To be Published
|
|
6B6L
 
 | | The crystal structure of glycosyl hydrolase family 2 (GH2) member from Bacteroides cellulosilyticus DSM 14838 | | Descriptor: | 1,2-ETHANEDIOL, FORMIC ACID, Glycosyl hydrolase family 2, ... | | Authors: | Tan, K, Joachimiak, G, Nocek, B, Enddres, M, Babnigg, G, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2017-10-02 | | Release date: | 2017-10-11 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | The crystal structure of glycosyl hydrolase family 2 (GH2) member from Bacteroides cellulosilyticus DSM 14838
To Be Published
|
|
6XPG
 
 | | Crystal Structure of Sialate O-acetylesterase from Bacteroides vulgatus by Serial Crystallography | | Descriptor: | Lysophospholipase L1 | | Authors: | Kim, Y, Sherrell, D.A, Owen, R, Axford, D, Ebrahim, A, Johnson, J, Welk, L, Babnigg, G, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2020-07-08 | | Release date: | 2020-07-15 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal Structure of Sialate O-acetylesterase from Bacteroides vulgatus by Serial Crystallography
To Be Published
|
|
6XPM
 
 | | Crystal Structure of Sialate O-acetylesterase from Bacteroides vulgatus with microfluidics crystals at room temperature | | Descriptor: | Lysophospholipase L1, SODIUM ION | | Authors: | Kim, Y, Johnson, J, Welk, L, Endres, M, Levens, A, Sherrell, D.A, Babnigg, G, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2020-07-08 | | Release date: | 2020-07-15 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal Structure of Sialate O-acetylesterase from Bacteroides vulgatus with microfluidics crystals at room temperature
To Be Published
|
|
3GKU
 
 | | Crystal structure of a probable RNA-binding protein from Clostridium symbiosum ATCC 14940 | | Descriptor: | Probable RNA-binding protein | | Authors: | Tan, K, Keigher, L, Jedrzejczak, R, Babnigg, G, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2009-03-11 | | Release date: | 2009-03-31 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.95 Å) | | Cite: | The crystal structure of a probable RNA-binding protein from Clostridium symbiosum ATCC 14940
To be Published
|
|
3GNJ
 
 | | The crystal structure of a thioredoxin-related protein from Desulfitobacterium hafniense DCB | | Descriptor: | Thioredoxin domain protein | | Authors: | Tan, K, Volkart, L, Gu, M, Kinney, J.N, Babnigg, G, Kerfeld, C, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2009-03-17 | | Release date: | 2009-05-12 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.99 Å) | | Cite: | The crystal structure of a thioredoxin-related protein from Desulfitobacterium hafniense DCB
To be Published
|
|
3HTR
 
 | | Crystal Structure of PRC-barrel Domain Protein from Rhodopseudomonas palustris | | Descriptor: | ACETIC ACID, ZINC ION, uncharacterized PRC-barrel Domain Protein | | Authors: | Kim, Y, Tesar, C, Jedrzejczak, R, Kinney, J, Babnigg, G, Harwood, C, Kerfeld, C, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2009-06-12 | | Release date: | 2009-07-07 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.06 Å) | | Cite: | Crystal Structure of PRC-barrel Domain Protein from Rhodopseudomonas palustris
To be Published
|
|
6UR7
 
 | | Crystal structure of Sel1 repeat protein from Oxalobacter formigenes | | Descriptor: | DI(HYDROXYETHYL)ETHER, GLYCEROL, SULFATE ION, ... | | Authors: | Chang, C, Tesar, C, Endres, M, Babnigg, G, Hassan, H, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2019-10-22 | | Release date: | 2020-10-28 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.709 Å) | | Cite: | Crystal structure of Sel1 repeat protein from Oxalobacter formigenes
To Be Published
|
|
6UUK
 
 | | Crystal structure of muramoyltetrapeptide carboxypeptidase from Oxalobacter formigenes | | Descriptor: | Muramoyltetrapeptide carboxypeptidase | | Authors: | Chang, C, Tesar, C, Endres, M, Babnigg, G, Hassan, H, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2019-10-30 | | Release date: | 2020-11-04 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.348 Å) | | Cite: | Crystal structure of muramoyltetrapeptide carboxypeptidase from Oxalobacter formigenes
To Be Published
|
|
6W9C
 
 | | The crystal structure of papain-like protease of SARS CoV-2 | | Descriptor: | CHLORIDE ION, Non-structural protein 3, ZINC ION | | Authors: | Osipiuk, J, Jedrzejczak, R, Tesar, C, Endres, M, Stols, L, Babnigg, G, Kim, Y, Michalska, K, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2020-03-22 | | Release date: | 2020-04-01 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | The crystal structure of papain-like protease of SARS CoV-2
to be published
|
|
4RGI
 
 | | Crystal Structure of KTSC Domain Protein YPO2434 from Yersinia pestis | | Descriptor: | GLYCEROL, SULFATE ION, Uncharacterized protein | | Authors: | Kim, Y, Chhor, G, Endres, M, Babnigg, G, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2014-09-30 | | Release date: | 2014-12-31 | | Method: | X-RAY DIFFRACTION (1.732 Å) | | Cite: | Crystal Structure of KTSC Domain Protein YPO2434 from Yersinia pestis
To be Published
|
|
4RGK
 
 | | Crystal Structure of Putative Phytanoyl-CoA Dioxygenase Family Protein YbiU from Yersinia pestis | | Descriptor: | 1,2-ETHANEDIOL, GLYCEROL, SULFATE ION, ... | | Authors: | Kim, Y, Chhor, G, Endres, M, Babnigg, G, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2014-09-30 | | Release date: | 2014-12-31 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Crystal Structure of Putative Phytanoyl-CoA Dioxygenase Family Protein YbiU from Yersinia pestis
To be Published
|
|
4RTF
 
 | | Crystal structure of molecular chaperone DnaK from Mycobacterium tuberculosis H37Rv | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, Chaperone protein DnaK, TETRAETHYLENE GLYCOL | | Authors: | Filippova, E.V, Minasov, G, Kiryukhina, O, Endres, M, Babnigg, G, Rubin, E, Sacchettini, J, Joachimiak, A, Anderson, W.F, Midwest Center for Structural Genomics (MCSG), Structures of Mtb Proteins Conferring Susceptibility to Known Mtb Inhibitors (MTBI) | | Deposit date: | 2014-11-14 | | Release date: | 2014-12-10 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.77 Å) | | Cite: | Crystal structure of molecular chaperone DnaK from Mycobacterium tuberculosis H37Rv
To be Published
|
|
4WKY
 
 | | Streptomcyes albus JA3453 oxazolomycin ketosynthase domain OzmN KS2 | | Descriptor: | 1,2-ETHANEDIOL, Beta-ketoacyl synthase, GLYCEROL, ... | | Authors: | Cuff, M.E, Mack, J.C, Endres, M, Babnigg, G, Bingman, C.A, Yennamalli, R, Lohman, J.R, Ma, M, Shen, B, Phillips Jr, G.N, Joachimiak, A, Midwest Center for Structural Genomics (MCSG), Enzyme Discovery for Natural Product Biosynthesis (NatPro) | | Deposit date: | 2014-10-03 | | Release date: | 2014-10-29 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural and evolutionary relationships of "AT-less" type I polyketide synthase ketosynthases.
Proc.Natl.Acad.Sci.USA, 112, 2015
|
|
4XR9
 
 | | Crystal structure of CalS8 from Micromonospora echinospora cocrystallized with NAD and TDP-glucose | | Descriptor: | CalS8, GLYCEROL, NICOTINAMIDE-ADENINE-DINUCLEOTIDE, ... | | Authors: | Michalska, K, Bigelow, L, Endres, M, Babnigg, G, Bingman, C.A, Yennamalli, R.M, Singh, S, Kharel, M.K, Thorson, J.S, Phillips Jr, G.N, Joachimiak, A, Midwest Center for Structural Genomics (MCSG), Enzyme Discovery for Natural Product Biosynthesis (NatPro) | | Deposit date: | 2015-01-20 | | Release date: | 2015-02-11 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Crystal structure of CalS8 from Micromonospora echinospora
To Be Published
|
|
4XRR
 
 | | Crystal structure of cals8 from micromonospora echinospora (P294S mutant) | | Descriptor: | CalS8, GLYCEROL | | Authors: | Michalska, K, Bigelow, L, Endres, M, Babnigg, G, Bingman, C.A, Yennamalli, R.M, Singh, S, Kharel, M.K, Thorson, J.S, Phillips Jr, G.N, Joachimiak, A, Midwest Center for Structural Genomics (MCSG), Enzyme Discovery for Natural Product Biosynthesis (NatPro) | | Deposit date: | 2015-01-21 | | Release date: | 2015-02-11 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.55 Å) | | Cite: | Structural Characterization of CalS8, a TDP-alpha-D-Glucose Dehydrogenase Involved in Calicheamicin Aminodideoxypentose Biosynthesis.
J. Biol. Chem., 290, 2015
|
|
6CKY
 
 | | Crystal structure of UcmS2 | | Descriptor: | Glyoxalase | | Authors: | Chang, C.Y, Chang, C, Annaval, T, Babnigg, G, Phillips Jr, G.N, Joachimiak, A, Shen, B. | | Deposit date: | 2018-03-01 | | Release date: | 2019-03-06 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal structure of UcmS2
To Be Published
|
|
