1CLP
 
 | | CRYSTAL STRUCTURE OF A CALCIUM-INDEPENDENT PHOSPHOLIPASELIKE MYOTOXIC PROTEIN FROM BOTHROPS ASPER VENOM | | Descriptor: | MYOTOXIN II | | Authors: | Arni, R.K, Ward, R.J, Gutierrez, J.M, Tulinsky, A. | | Deposit date: | 1994-09-12 | | Release date: | 1994-11-30 | | Last modified: | 2024-06-05 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structure of a calcium-independent phospholipase-like myotoxic protein from Bothrops asper venom.
Acta Crystallogr.,Sect.D, 51, 1995
|
|
1GOD
 
 | | MONOMERIC LYS-49 PHOSPHOLIPASE A2 HOMOLOGUE ISOLATED FROM THE VENOM OF CERROPHIDION (BOTHROPS) GODMANI | | Descriptor: | PROTEIN (PHOSPHOLIPASE A2) | | Authors: | Arni, R.K, Fontes, M.R.M, Barberato, C, Gutierrez, J.M, Diaz-Oreiro, C, Ward, R.J. | | Deposit date: | 1999-04-16 | | Release date: | 1999-04-23 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Crystal structure of myotoxin II, a monomeric Lys49-phospholipase A2 homologue isolated from the venom of Cerrophidion (Bothrops) godmani.
Arch.Biochem.Biophys., 366, 1999
|
|
1B2M
 
 | | THREE-DIMENSIONAL STRUCTURE OF RIBONULCEASE T1 COMPLEXED WITH AN ISOSTERIC PHOSPHONATE ANALOGUE OF GPU: ALTERNATE SUBSTRATE BINDING MODES AND CATALYSIS. | | Descriptor: | 5'-R(*GP*(U34))-3', RIBONUCLEASE T1 | | Authors: | Arni, R.K, Watanabe, L, Ward, R.J, Kreitman, R.J, Kumar, K, Walz Jr, F.G. | | Deposit date: | 1998-11-27 | | Release date: | 1999-03-25 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Three-dimensional structure of ribonuclease T1 complexed with an isosteric phosphonate substrate analogue of GpU: alternate substrate binding modes and catalysis.
Biochemistry, 38, 1999
|
|
1RN1
 
 | | THREE-DIMENSIONAL STRUCTURE OF GLN 25-RIBONUCLEASE T1 AT 1.84 ANGSTROMS RESOLUTION: STRUCTURAL VARIATIONS AT THE BASE RECOGNITION AND CATALYTIC SITES | | Descriptor: | RIBONUCLEASE T1 ISOZYME, SULFATE ION | | Authors: | Arni, R.K, Pal, G.P, Ravichandran, K.G, Tulinsky, A, Walz Junior, F.G, Metcalf, P. | | Deposit date: | 1991-11-22 | | Release date: | 1994-01-31 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.84 Å) | | Cite: | Three-dimensional structure of Gln25-ribonuclease T1 at 1.84-A resolution: structural variations at the base recognition and catalytic sites.
Biochemistry, 31, 1992
|
|
1XXS
 
 | | Structural insights for fatty acid binding in a Lys49 phospholipase A2: crystal structure of myotoxin II from Bothrops moojeni complexed with stearic acid | | Descriptor: | Phospholipase A2 homolog 2, STEARIC ACID, SULFATE ION | | Authors: | Watanabe, L, Soares, A.M, Ward, R.J, Fontes, M.R, Arni, R.K. | | Deposit date: | 2004-11-08 | | Release date: | 2005-03-29 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structural insights for fatty acid binding in a Lys49-phospholipase A(2): crystal structure of myotoxin II from Bothrops moojeni complexed with stearic acid
Biochimie, 87, 2005
|
|
2PH4
 
 | | Crystal structure of a novel Arg49 phospholipase A2 homologue from Zhaoermia mangshanensis venom | | Descriptor: | DI(HYDROXYETHYL)ETHER, SULFATE ION, Zhaoermiatoxin | | Authors: | Murakami, M.T, Kuch, U, Mebs, D, Arni, R.K. | | Deposit date: | 2007-04-10 | | Release date: | 2008-03-18 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Crystal structure of a novel myotoxic Arg49 phospholipase A(2) homolog (zhaoermiatoxin) from Zhaoermia mangshanensis snake venom: Insights into Arg49 coordination and the role of Lys122 in the polarization of the C-terminus.
Toxicon, 51, 2008
|
|
5O6F
 
 | | NMR structure of cold shock protein A from Corynebacterium pseudotuberculosis | | Descriptor: | Cold-shock protein | | Authors: | Caruso, I.P, Panwalkar, V, Coronado, M.A, Dingley, A.J, Cornelio, M.L, Willbold, D, Arni, R.K, Eberle, R.J. | | Deposit date: | 2017-06-06 | | Release date: | 2017-07-19 | | Last modified: | 2024-06-19 | | Method: | SOLUTION NMR | | Cite: | Structure and interaction of Corynebacterium pseudotuberculosis cold shock protein A with Y-box single-stranded DNA fragment.
FEBS J., 285, 2018
|
|
1ZZZ
 
 | | Trypsin inhibitors with rigid tripeptidyl aldehydes | | Descriptor: | 2-{(3S)-3-[(benzylsulfonyl)amino]-2-oxopiperidin-1-yl}-N-{(2S)-1-[(3R)-1-carbamimidoylpiperidin-3-yl]-3-oxopropan-2-yl}acetamide, CALCIUM ION, TRYPSIN | | Authors: | Krishnan, R, Zhang, E, Hakansson, K, Arni, R.K, Tulinsky, A, Lim-Wilby, M.S.L, Levy, O.E, Semple, J.E, Brunck, T.K. | | Deposit date: | 1998-06-02 | | Release date: | 1999-06-15 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Highly selective mechanism-based thrombin inhibitors: structures of thrombin and trypsin inhibited with rigid peptidyl aldehydes.
Biochemistry, 37, 1998
|
|
8T3J
 
 | | Crystal structure of native exfoliative toxin C (ExhC) from Mammaliicoccus sciuri | | Descriptor: | Exfoliative toxin C | | Authors: | Calil, F.A, Gismene, C, Hernandez Gonzalez, J.E, Ziem Nascimento, A.F, Santisteban, A.R.N, Arni, R.K, Barros Mariutti, R. | | Deposit date: | 2023-06-07 | | Release date: | 2023-11-08 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.705 Å) | | Cite: | Necrotic activity of ExhC from Mammaliicoccus sciuri is mediated by specific amino acid residues.
Int.J.Biol.Macromol., 254, 2024
|
|
8T3I
 
 | | Crystal structure of mutant exfoliative toxin C (ExhC) from Mammaliicoccus sciuri | | Descriptor: | Exfoliative toxin C | | Authors: | Gismene, C, Calil, F.A, Hernandez Gonzalez, J.E, Ziem Nascimento, A.F, Santisteban, A.R.N, Arni, R.K, Barros Mariutti, R. | | Deposit date: | 2023-06-07 | | Release date: | 2023-11-08 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.57 Å) | | Cite: | Necrotic activity of ExhC from Mammaliicoccus sciuri is mediated by specific amino acid residues.
Int.J.Biol.Macromol., 254, 2024
|
|
8DAX
 
 | | New insights into the P186 flip and oligomeric state of Staphylococcus aureus exfoliative toxin E: implications for the exfoliative mechanism | | Descriptor: | Exfoliative toxin E | | Authors: | Gismene, C, Nascimento, A.F.Z, Hernandez-Gonzalez, J.E, Santisteban, A.R.N, de Moraes, F.R, Arni, R.K, Mariutti, R.B. | | Deposit date: | 2022-06-14 | | Release date: | 2022-11-02 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.61 Å) | | Cite: | Staphylococcus aureus Exfoliative Toxin E, Oligomeric State and Flip of P186: Implications for Its Action Mechanism.
Int J Mol Sci, 23, 2022
|
|
3UX7
 
 | | Crystal structure of a dimeric myotoxic component of the Vipera ammodytes meridionalis venom reveals determinants of myotoxicity and membrane damaging activity | | Descriptor: | Ammodytin L(1) isoform, SULFATE ION | | Authors: | Georgieva, D, Coronado, M, Oberthuer, D, Buck, F, Duhalov, D, Arni, R.K, Genov, N, Betzel, C. | | Deposit date: | 2011-12-04 | | Release date: | 2012-04-25 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.97 Å) | | Cite: | Crystal structure of a dimeric Ser49 PLA(2)-like myotoxic component of the Vipera ammodytes meridionalis venomics reveals determinants of myotoxicity and membrane damaging activity.
Mol Biosyst, 8, 2012
|
|
1ZLB
 
 | | Crystal structure of catalytically-active phospholipase A2 in the absence of calcium | | Descriptor: | hypotensive phospholipase A2 | | Authors: | Murakami, M.T, Cintra, A.C, Gabdoulkhakov, A, Genov, N, Betzel, C, Arni, R.K. | | Deposit date: | 2005-05-05 | | Release date: | 2006-04-25 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (0.97 Å) | | Cite: | Insights into metal ion binding in phospholipases A(2): ultra high-resolution crystal structures of an acidic phospholipase A(2) in the Ca(2+) free and bound states.
Biochimie, 88, 2006
|
|
1XX1
 
 | | Structural basis for ion-coordination and the catalytic mechanism of sphingomyelinases D | | Descriptor: | 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, MAGNESIUM ION, SULFATE ION, ... | | Authors: | Murakami, M.T, Tambourgi, D.V, Arni, R.K. | | Deposit date: | 2004-11-03 | | Release date: | 2005-01-18 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Structural basis for metal ion coordination and the catalytic mechanism of sphingomyelinases d
J.Biol.Chem., 280, 2005
|
|
1ZL7
 
 | | Crystal structure of catalytically-active phospholipase A2 with bound calcium | | Descriptor: | CALCIUM ION, GLYCEROL, hypotensive phospholipase A2 | | Authors: | Murakami, M.T, Cintra, A.C, Gabdoulkhakov, A, Genov, N, Betzel, C, Arni, R.K. | | Deposit date: | 2005-05-05 | | Release date: | 2006-04-25 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Insights into metal ion binding in phospholipases A(2): ultra high-resolution crystal structures of an acidic phospholipase A(2) in the Ca(2+) free and bound states.
Biochimie, 88, 2006
|
|
5JVO
 
 | | Crystal structure of the Arginine Repressor from the pathogenic bacterium Corynebacterium pseudotuberculosis | | Descriptor: | Arginine repressor, SULFATE ION, TYROSINE | | Authors: | Mariutti, R.B, Ullah, A, Murakami, M.T, Arni, R.K. | | Deposit date: | 2016-05-11 | | Release date: | 2016-08-31 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Tyrosine binding and promiscuity in the arginine repressor from the pathogenic bacterium Corynebacterium pseudotuberculosis.
Biochem.Biophys.Res.Commun., 475, 2016
|
|
4RW3
 
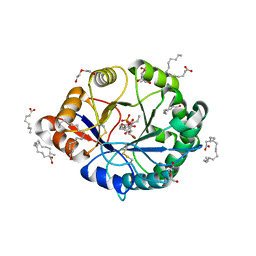 | | Structural insights into substrate binding of brown spider venom class II phospholipases D | | Descriptor: | D-MYO-INOSITOL-1-PHOSPHATE, DECANOIC ACID, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Coronado, M.A, Ullah, A, da Silva, L.S, Chaves-Moreira, D, Vuitika, L, Chaim, O.M, Veiga, S.S, Chahine, J, Murakami, M.T, Arni, R.K. | | Deposit date: | 2014-12-01 | | Release date: | 2015-06-03 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.72 Å) | | Cite: | Structural Insights into Substrate Binding of Brown Spider Venom Class II Phospholipases D.
Curr Protein Pept Sci, 16, 2015
|
|
4RW5
 
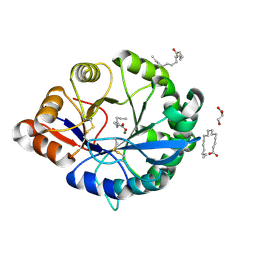 | | Structural insights into substrate binding of brown spider venom class II phospholipases D | | Descriptor: | GLYCEROL, MAGNESIUM ION, N-TRIDECANOIC ACID, ... | | Authors: | Coronado, M.A, Ullah, A, da Silva, L.S, Chaves-Moreira, D, Vuitika, L, Chaim, O.M, Veiga, S.S, Chahine, J, Murakami, M.T, Arni, R.K. | | Deposit date: | 2014-12-01 | | Release date: | 2015-06-03 | | Last modified: | 2015-12-16 | | Method: | X-RAY DIFFRACTION (1.64 Å) | | Cite: | Structural Insights into Substrate Binding of Brown Spider Venom Class II Phospholipases D.
Curr Protein Pept Sci, 16, 2015
|
|
6WJP
 
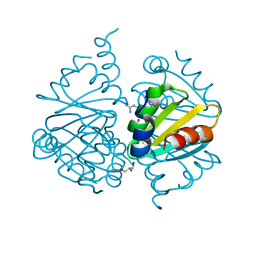 | | Crystal structure of Arginine Repressor P115Q mutant from the pathogenic bacterium Corynebacterium pseudotuberculosis bound to arginine | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, ACETATE ION, ARGININE, ... | | Authors: | Nascimento, A.F.Z, Hernandez-Gonzalez, J.E, de Morais, M.A.B, Murakami, M.T, Carareto, C.M.A, Arni, R.K, Mariutti, R.B. | | Deposit date: | 2020-04-14 | | Release date: | 2020-04-22 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.701 Å) | | Cite: | A single P115Q mutation modulates specificity in the Corynebacterium pseudotuberculosis arginine repressor.
Biochim Biophys Acta Gen Subj, 1864, 2020
|
|
6WJO
 
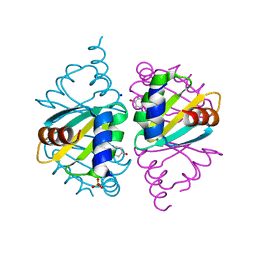 | | Crystal structure of wild-type Arginine Repressor from the pathogenic bacterium Corynebacterium pseudotuberculosis bound to tyrosine | | Descriptor: | Arginine repressor, SODIUM ION, SULFATE ION, ... | | Authors: | Nascimento, A.F.Z, Hernandez-Gonzalez, J.E, de Morais, M.A.B, Murakami, M.T, Carareto, C.M.A, Arni, R.K, Mariutti, R.B. | | Deposit date: | 2020-04-14 | | Release date: | 2020-04-22 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.693 Å) | | Cite: | A single P115Q mutation modulates specificity in the Corynebacterium pseudotuberculosis arginine repressor.
Biochim Biophys Acta Gen Subj, 1864, 2020
|
|
1KI0
 
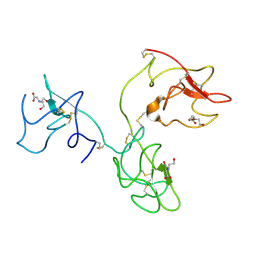 | | The X-ray Structure of Human Angiostatin | | Descriptor: | ANGIOSTATIN, BICINE | | Authors: | Abad, M.C, Arni, R.K, Grella, D.K, Castellino, F.J, Tulinsky, A, Geiger, J.H. | | Deposit date: | 2001-12-02 | | Release date: | 2002-05-29 | | Last modified: | 2021-10-27 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | The X-ray crystallographic structure of the angiogenesis inhibitor angiostatin.
J.Mol.Biol., 318, 2002
|
|
2AIP
 
 | |
2AIQ
 
 | |
2AOZ
 
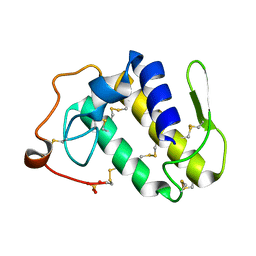 | | Crystal structure of the myotoxin-II from Atropoides nummifer venom | | Descriptor: | Phospholipase A2 homolog, SULFATE ION | | Authors: | Melo, C.C, Murakami, M.T, Angulo, Y, Lomonte, B, Arni, R.K. | | Deposit date: | 2005-08-15 | | Release date: | 2006-07-25 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.08 Å) | | Cite: | Structure of myotoxin II, a catalytically inactive Lys49 phospholipase A2 homologue from Atropoides nummifer venom.
Acta Crystallogr.,Sect.F, 62, 2006
|
|
4DCF
 
 | | Structure of MTX-II from Bothrops brazili | | Descriptor: | MTX-II chain A, TETRAETHYLENE GLYCOL | | Authors: | Ullah, A, Souza, T.A.C.B, Betzel, C, Murakami, M.T, Arni, R.K. | | Deposit date: | 2012-01-17 | | Release date: | 2012-06-13 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Crystallographic portrayal of different conformational states of a Lys49 phospholipase A2 homologue: insights into structural determinants for myotoxicity and dimeric configuration.
Int.J.Biol.Macromol., 51, 2012
|
|
