7WEM
 
 | | Solid-state NMR Structure of TFo c-Subunit Ring | | Descriptor: | ATP synthase subunit c | | Authors: | Akutsu, H, Todokoro, Y, Kang, S.-J, Suzuki, T, Yoshida, M, Ikegami, T, Fujiwara, T. | | Deposit date: | 2021-12-23 | | Release date: | 2022-08-10 | | Last modified: | 2024-05-15 | | Method: | SOLID-STATE NMR | | Cite: | Chemical Conformation of the Essential Glutamate Site of the c -Ring within Thermophilic Bacillus F o F 1 -ATP Synthase Determined by Solid-State NMR Based on its Isolated c -Ring Structure.
J.Am.Chem.Soc., 144, 2022
|
|
1SNH
 
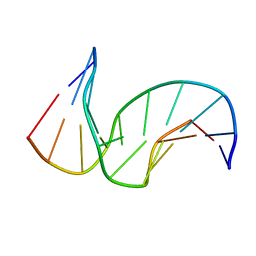 | | Solution structure of the DNA Decamer Duplex Containing Double TG Mismatches of Cis-syn Cyclobutane Pyrimidine Dimer | | Descriptor: | 5'-D(*CP*GP*CP*AP*TP*TP*AP*CP*GP*C)-3', 5'-D(*GP*CP*GP*TP*GP*GP*TP*GP*CP*G)-3' | | Authors: | Lee, J.H, Park, C.J, Shin, J.S, Ikegami, T, Akutsu, H, Choi, B.S. | | Deposit date: | 2004-03-11 | | Release date: | 2004-05-18 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | NMR structure of the DNA decamer duplex containing double T*G mismatches of cis-syn cyclobutane pyrimidine dimer: implications for DNA damage recognition by the XPC-hHR23B complex.
Nucleic Acids Res., 32, 2004
|
|
8XZ2
 
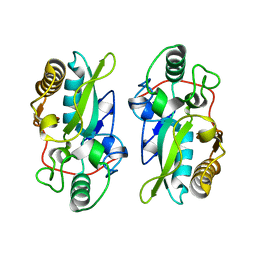 | | The structural model of a homodimeric D-Ala-D-Ala metallopeptidase, VanX, from vancomycin-resistant bacteria | | Descriptor: | D-alanyl-D-alanine dipeptidase | | Authors: | Konuma, T, Takai, T, Tsuchiya, C, Nishida, M, Hashiba, M, Yamada, Y, Shirai, H, Motoda, Y, Nagadoi, A, Chikaishi, E, Akagi, K, Akashi, S, Yamazaki, T, Akutsu, H, Oe, A, Ikegami, T. | | Deposit date: | 2024-01-20 | | Release date: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Analysis of the homodimeric structure of a D-Ala-D-Ala metallopeptidase, VanX, from vancomycin-resistant bacteria.
Protein Sci., 33, 2024
|
|
1WUZ
 
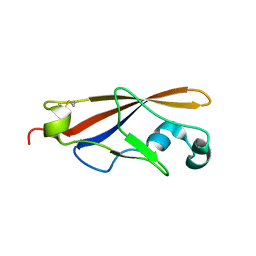 | | Structure of EC1 domain of CNR | | Descriptor: | Pcdha4 protein | | Authors: | Morishita, H, Umitsu, M, Yamaguchi, T, Murata, Y, Shibata, N, Udaka, K, Higuchi, Y, Akutsu, H, Yagi, T, Ikegami, T. | | Deposit date: | 2004-12-09 | | Release date: | 2005-12-13 | | Last modified: | 2024-10-09 | | Method: | SOLUTION NMR | | Cite: | Structural diversity of the first cadherin domains revealed by the structure of CNR/Protocadherin alpha
To be Published
|
|
1J0O
 
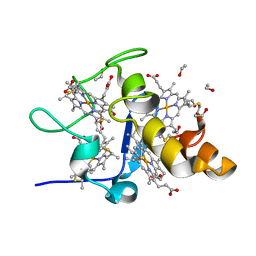 | | High Resolution Crystal Structure of the wild type Tetraheme Cytochrome c3 from Desulfovibrio vulgaris Miyazaki F | | Descriptor: | Cytochrome c3, ETHANOL, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Ozawa, K, Yasukawa, F, Kumagai, J, Ohmura, T, Cusanvich, M.A, Tomimoto, Y, Ogata, H, Higuchi, Y, Akutsu, H. | | Deposit date: | 2002-11-19 | | Release date: | 2003-11-19 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.15 Å) | | Cite: | Role of the aromatic ring of Tyr43 in tetraheme cytochrome c(3) from Desulfovibrio vulgaris Miyazaki F.
Biophys.J., 85, 2003
|
|
1J0P
 
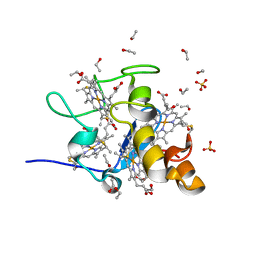 | | Three dimensional Structure of the Y43L mutant of Tetraheme Cytochrome c3 from Desulfovibrio vulgaris Miyazaki F | | Descriptor: | Cytochrome c3, ETHANOL, PROTOPORPHYRIN IX CONTAINING FE, ... | | Authors: | Ozawa, K, Yasukawa, F, Kumagai, J, Ohmura, T, Cusanvich, M.A, Tomimoto, Y, Ogata, H, Higuchi, Y, Akutsu, H. | | Deposit date: | 2002-11-19 | | Release date: | 2003-11-19 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (0.91 Å) | | Cite: | Role of the aromatic ring of Tyr43 in tetraheme cytochrome c(3) from Desulfovibrio vulgaris Miyazaki F.
Biophys.J., 85, 2003
|
|
1WU0
 
 | |
2E5U
 
 | |
2E5Y
 
 | |
2E5T
 
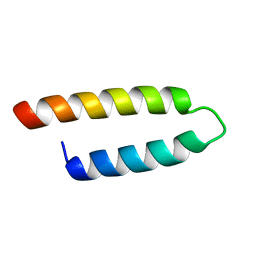 | |
1IT1
 
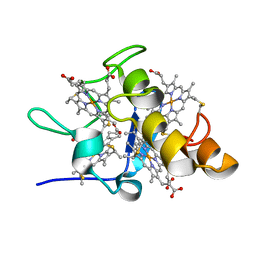 | | Solution structures of ferrocytochrome c3 from Desulfovibrio vulgaris Miyazaki F | | Descriptor: | HEME C, cytochrome c3 | | Authors: | Harada, E, Fukuoka, Y, Ohmura, T, Fukunishi, A, Kawai, G, Fujiwara, T, Akutsu, H. | | Deposit date: | 2001-12-29 | | Release date: | 2002-07-10 | | Last modified: | 2024-11-13 | | Method: | SOLUTION NMR | | Cite: | Redox-coupled conformational alternations in cytochrome c(3) from D. vulgaris Miyazaki F on the basis of its reduced solution structure.
J.Mol.Biol., 319, 2002
|
|
2D49
 
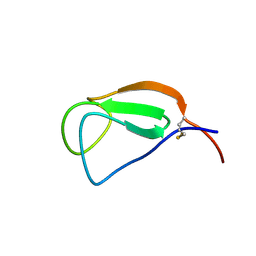 | | Solution structure of the Chitin-Binding Domain of Streptomyces griseus Chitinase C | | Descriptor: | chitinase C | | Authors: | Akagi, K, Watanabe, J, Hara, M, Kezuka, Y, Chikaishi, E, Yamaguchi, T, Akutsu, H, Nonaka, T, Watanabe, T, Ikegami, T. | | Deposit date: | 2005-10-11 | | Release date: | 2006-10-11 | | Last modified: | 2024-10-23 | | Method: | SOLUTION NMR | | Cite: | Identification of the substrate interaction region of the chitin-binding domain of Streptomyces griseus chitinase C
J.Biochem.(Tokyo), 139, 2006
|
|
2CVC
 
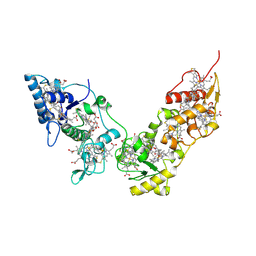 | | Crystal structure of High-Molecular Weight Cytochrome c from Desulfovibrio vulgaris (Hildenborough) | | Descriptor: | HEME C, High-molecular-weight cytochrome c precursor | | Authors: | Suto, K, Sato, M, Shibata, N, Kitamura, M, Morimoto, Y, Takayama, Y, Ozawa, K, Akutsu, H, Higuchi, Y, Yasuoka, N. | | Deposit date: | 2005-06-02 | | Release date: | 2006-06-06 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structure of High-Molecular Weight Cytochrome c
To be Published
|
|
2CZP
 
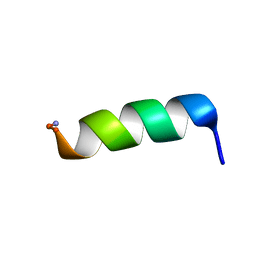 | | Structural analysis of membrane-bound mastoparan-X by solid-state NMR | | Descriptor: | Mastoparan X | | Authors: | Todokoro, Y, Fujiwara, T, Yumen, I, Fukushima, K, Kang, S.-W, Park, J.-S, Kohno, T, Wakamatsu, K, Akutsu, H. | | Deposit date: | 2005-07-14 | | Release date: | 2006-07-04 | | Last modified: | 2024-11-13 | | Method: | SOLID-STATE NMR | | Cite: | Structure of tightly membrane-bound mastoparan-x, a g-protein-activating Peptide, determined by solid-state NMR.
Biophys.J., 91, 2006
|
|
2RQ7
 
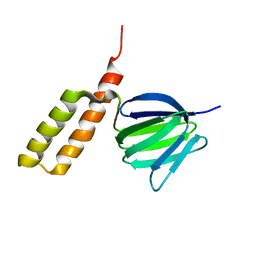 | | Solution structure of the epsilon subunit chimera combining the N-terminal beta-sandwich domain from T. Elongatus bp-1 f1 and the C-terminal alpha-helical domain from spinach chloroplast F1 | | Descriptor: | ATP synthase epsilon chain,ATP synthase epsilon chain, chloroplastic | | Authors: | Yagi, H, Konno, H, Murakami-Fuse, T, Oroguchi, H, Akutsu, T, Ikeguchi, M, Hisabori, T. | | Deposit date: | 2009-03-03 | | Release date: | 2010-01-12 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Structural and functional analysis of the intrinsic inhibitor subunit epsilon of F1-ATPase from photosynthetic organisms.
Biochem.J., 425, 2010
|
|
2RQ6
 
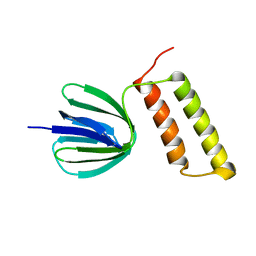 | | Solution structure of the epsilon subunit of the F1-atpase from thermosynechococcus elongatus BP-1 | | Descriptor: | ATP synthase epsilon chain | | Authors: | Yagi, H, Konno, H, Murakami-Fuse, T, Oroguchi, H, Akutsu, T, Ikeguchi, M, Hisabori, T. | | Deposit date: | 2009-03-03 | | Release date: | 2010-01-12 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Structural and functional analysis of the intrinsic inhibitor subunit epsilon of F1-ATPase from photosynthetic organisms.
Biochem.J., 425, 2010
|
|
2YYW
 
 | |
2YYX
 
 | |
2YXC
 
 | |
2Z47
 
 | |
1WR5
 
 | |
1PIB
 
 | | Solution structure of DNA containing CPD opposited by GA | | Descriptor: | 5'-D(*CP*GP*CP*AP*TP*TP*AP*CP*GP*C)-3', 5'-D(*GP*CP*GP*TP*GP*AP*TP*GP*CP*G)-3' | | Authors: | Lee, J.H, Park, C.J, Shin, J.S, Choi, B.S. | | Deposit date: | 2003-05-30 | | Release date: | 2004-05-18 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | NMR structure of the DNA decamer duplex containing double T*G mismatches of cis-syn cyclobutane pyrimidine dimer: implications for DNA damage recognition by the XPC-hHR23B complex.
Nucleic Acids Res., 32, 2004
|
|
2FFN
 
 | |
2EWK
 
 | |
2E84
 
 | | Crystal structure of High-Molecular Weight Cytochrome c from Desulfovibrio vulgaris (Miyazaki F) in the presence of zinc ion | | Descriptor: | High-molecular-weight cytochrome c, PROTOPORPHYRIN IX CONTAINING FE, SODIUM ION, ... | | Authors: | Shibata, N, Suto, K, Sato, M, Morimoto, Y, Kitamura, M, Higuchi, Y. | | Deposit date: | 2007-01-17 | | Release date: | 2008-01-15 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Structure of High-Molecular Weight Cytochrome c from Desulfovibrio vulgaris (Miyazaki F)
To be Published
|
|
