8BNW
 
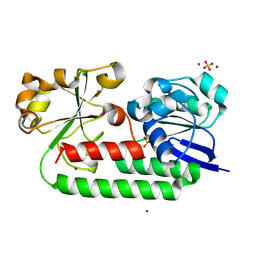 | | X-ray structure of the CeuE Homologue from Parageobacillus thermoglucosidasius - apo form | | Descriptor: | ABC transporter, NICKEL (II) ION, SULFATE ION | | Authors: | Blagova, E.V, Bennett, M, Booth, R, Dodson, E.J, Duhme-KLair, A.-K, Wilson, K.S. | | Deposit date: | 2022-11-14 | | Release date: | 2023-07-12 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.133 Å) | | Cite: | Thermostable homologues of the periplasmic siderophore-binding protein CeuE from Geobacillus stearothermophilus and Parageobacillus thermoglucosidasius.
Acta Crystallogr D Struct Biol, 79, 2023
|
|
2IGD
 
 | |
2JEQ
 
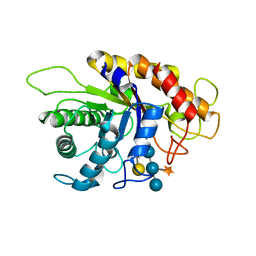 | | Family 5 xyloglucanase from Paenibacillus pabuli in complex with ligand | | Descriptor: | XYLOGLUCANASE, beta-D-glucopyranose-(1-4)-[alpha-D-xylopyranose-(1-6)]beta-D-glucopyranose-(1-4)-[beta-D-galactopyranose-(1-2)-alpha-D-xylopyranose-(1-6)]beta-D-glucopyranose-(1-4)-beta-D-glucopyranose | | Authors: | Gloster, T.M, Ibatullin, F.M, Macauley, K, Eklof, J.M, Roberts, S, Turkenburg, J.P, Bjornvad, M.E, Jorgensen, P.L, Danielsen, S, Johansen, K, Borchert, T.V, Wilson, K.S, Brumer, H, Davies, G.J. | | Deposit date: | 2007-01-18 | | Release date: | 2007-03-20 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.94 Å) | | Cite: | Characterization and Three-Dimensional Structures of Two Distinct Bacterial Xyloglucanases from Families Gh5 and Gh12.
J.Biol.Chem., 282, 2007
|
|
2J8W
 
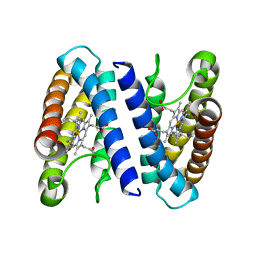 | | The crystal structure of cytochrome c' from Rubrivivax gelatinosus at 1.3 A Resolution and pH 8.0 | | Descriptor: | CYTOCHROME C', HEME C | | Authors: | Benini, S, Ciurli, S, Rypniewski, W.R, Wilson, K.S. | | Deposit date: | 2006-10-30 | | Release date: | 2007-11-06 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.29 Å) | | Cite: | High resolution crystal structure of Rubrivivax gelatinosus cytochrome c'.
J. Inorg. Biochem., 102, 2008
|
|
2JEP
 
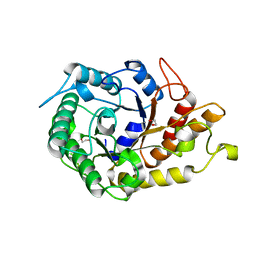 | | Native family 5 xyloglucanase from Paenibacillus pabuli | | Descriptor: | 1,2-ETHANEDIOL, CALCIUM ION, XYLOGLUCANASE | | Authors: | Gloster, T.M, Ibatullin, F.M, Macauley, K, Eklof, J.M, Roberts, S, Turkenburg, J.P, Bjornvad, M.E, Jorgensen, P.L, Danielsen, S, Johansen, K, Borchert, T.V, Wilson, K.S, Brumer, H, Davies, G.J. | | Deposit date: | 2007-01-18 | | Release date: | 2007-03-20 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Characterization and Three-Dimensional Structures of Two Distinct Bacterial Xyloglucanases from Families Gh5 and Gh12.
J.Biol.Chem., 282, 2007
|
|
2JHF
 
 | | Structural evidence for a ligand coordination switch in liver alcohol dehydrogenase | | Descriptor: | ALCOHOL DEHYDROGENASE E CHAIN, CADMIUM ION, DIMETHYL SULFOXIDE, ... | | Authors: | Meijers, R, Adolph, H.W, Dauter, Z, Wilson, K.S, Lamzin, V.S, Cedergren-Zeppezauer, E.S. | | Deposit date: | 2007-02-22 | | Release date: | 2007-04-24 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1 Å) | | Cite: | Structural Evidence for a Ligand Coordination Switch in Liver Alcohol Dehydrogenase
Biochemistry, 46, 2007
|
|
2JHG
 
 | | Structural evidence for a ligand coordination switch in liver alcohol dehydrogenase | | Descriptor: | 2-METHYLPROPANAMIDE, ALCOHOL DEHYDROGENASE E CHAIN, NICOTINAMIDE-ADENINE-DINUCLEOTIDE, ... | | Authors: | Meijers, R, Adolph, H.W, Dauter, Z, Wilson, K.S, Lamzin, V.S, Cedergren-Zeppezauer, E.S. | | Deposit date: | 2007-02-22 | | Release date: | 2007-04-24 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.2 Å) | | Cite: | Structural Evidence for a Ligand Coordination Switch in Liver Alcohol Dehydrogenase
Biochemistry, 46, 2007
|
|
1HUE
 
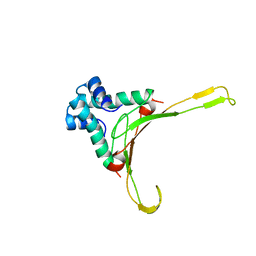 | | HISTONE-LIKE PROTEIN | | Descriptor: | HU PROTEIN | | Authors: | Vis, H, Mariani, M, Vorgias, C.E, Wilson, K.S, Kaptein, R, Boelens, R. | | Deposit date: | 1995-05-26 | | Release date: | 1995-10-15 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the HU protein from Bacillus stearothermophilus.
J.Mol.Biol., 254, 1995
|
|
1KF2
 
 | | Atomic Resolution Structure of RNase A at pH 5.2 | | Descriptor: | SULFATE ION, pancreatic ribonuclease | | Authors: | Berisio, R, Sica, F, Lamzin, V.S, Wilson, K.S, Zagari, A, Mazzarella, L. | | Deposit date: | 2001-11-19 | | Release date: | 2001-12-19 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (1.1 Å) | | Cite: | Atomic resolution structures of ribonuclease A at six pH values.
Acta Crystallogr.,Sect.D, 58, 2002
|
|
1KF4
 
 | | Atomic Resolution Structure of RNase A at pH 6.3 | | Descriptor: | SULFATE ION, pancreatic ribonuclease | | Authors: | Berisio, R, Sica, F, Lamzin, V.S, Wilson, K.S, Zagari, A, Mazzarella, L. | | Deposit date: | 2001-11-19 | | Release date: | 2001-12-19 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (1.1 Å) | | Cite: | Atomic resolution structures of ribonuclease A at six pH values.
Acta Crystallogr.,Sect.D, 58, 2002
|
|
1KF7
 
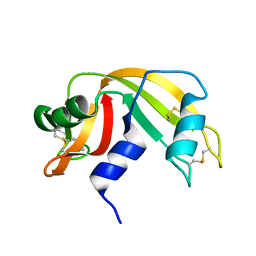 | | Atomic Resolution Structure of RNase A at pH 8.0 | | Descriptor: | pancreatic ribonuclease | | Authors: | Berisio, R, Sica, F, Lamzin, V.S, Wilson, K.S, Zagari, A, Mazzarella, L. | | Deposit date: | 2001-11-19 | | Release date: | 2001-12-19 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (1.15 Å) | | Cite: | Atomic resolution structures of ribonuclease A at six pH values.
Acta Crystallogr.,Sect.D, 58, 2002
|
|
1KF8
 
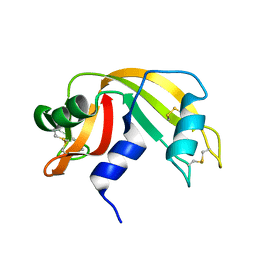 | | Atomic resolution structure of RNase A at pH 8.8 | | Descriptor: | pancreatic ribonuclease | | Authors: | Berisio, R, Sica, F, Lamzin, V.S, Wilson, K.S, Zagari, A, Mazzarella, L. | | Deposit date: | 2001-11-19 | | Release date: | 2001-12-19 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (1.15 Å) | | Cite: | Atomic resolution structures of ribonuclease A at six pH values.
Acta Crystallogr.,Sect.D, 58, 2002
|
|
1KF3
 
 | | Atomic Resolution Structure of RNase A at pH 5.9 | | Descriptor: | SULFATE ION, pancreatic ribonuclease | | Authors: | Berisio, R, Sica, F, Lamzin, V.S, Wilson, K.S, Zagari, A, Mazzarella, L. | | Deposit date: | 2001-11-19 | | Release date: | 2001-12-19 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (1.05 Å) | | Cite: | Atomic resolution structures of ribonuclease A at six pH values.
Acta Crystallogr.,Sect.D, 58, 2002
|
|
1KF5
 
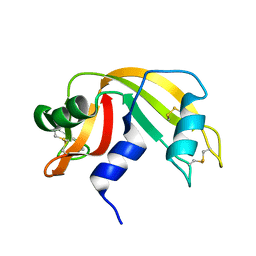 | | Atomic Resolution Structure of RNase A at pH 7.1 | | Descriptor: | pancreatic ribonuclease | | Authors: | Berisio, R, Sica, F, Lamzin, V.S, Wilson, K.S, Zagari, A, Mazzarella, L. | | Deposit date: | 2001-11-19 | | Release date: | 2001-12-19 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (1.15 Å) | | Cite: | Atomic resolution structures of ribonuclease A at six pH values.
Acta Crystallogr.,Sect.D, 58, 2002
|
|
3DD6
 
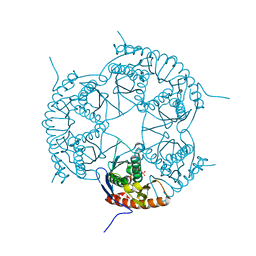 | | Crystal structure of Rph, an exoribonuclease from Bacillus anthracis at 1.7 A resolution | | Descriptor: | Ribonuclease PH, SULFATE ION | | Authors: | Rawlings, A.E, Blagova, E.V, Levdikov, V.M, Fogg, M.J, Wilson, K.S, Wilkinson, A.J, Structural Proteomics in Europe 2 (SPINE-2) | | Deposit date: | 2008-06-05 | | Release date: | 2009-02-10 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.702 Å) | | Cite: | The structure of Rph, an exoribonuclease from Bacillus anthracis, at 1.7 A resolution.
Acta Crystallogr.,Sect.F, 65, 2009
|
|
3T6Y
 
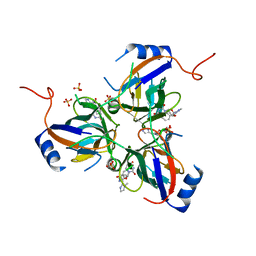 | | 5'-Diphenyl Nucleoside Inhibitors of Plasmodium falciparum dUTPase | | Descriptor: | 2',5'-dideoxy-5'-{[(R)-(1-methyl-1H-imidazol-2-yl)(phenyl)methyl]amino}uridine, Deoxyuridine 5'-triphosphate nucleotidohydrolase, putative, ... | | Authors: | Hampton, S.E, Baragana, B, Schipani, A, Bosch-Navarrete, C, Musso-Buendia, A, Recio, E, Kaiser, M, Whittingham, J.L, Roberts, S.M, Shevtsov, M, Brannigan, J.A, Kahnberg, P, Brun, R, Wilson, K.S, Gonzalez-Pacanowska, D, Johansson, N.G, Gilbert, I.H. | | Deposit date: | 2011-07-29 | | Release date: | 2012-08-29 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Design, synthesis, and evaluation of 5'-diphenyl nucleoside analogues as inhibitors of the Plasmodium falciparum dUTPase.
Chemmedchem, 6, 2011
|
|
4OYY
 
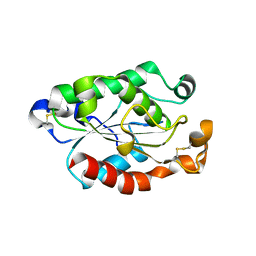 | |
3T60
 
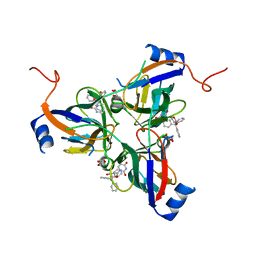 | | 5'-Diphenyl Nucleoside Inhibitors of Plasmodium falciparum dUTPase | | Descriptor: | 2',5'-dideoxy-5'-(tritylamino)uridine, Deoxyuridine 5'-triphosphate nucleotidohydrolase, putative, ... | | Authors: | Hampton, S.E, Baragana, B, Schipani, A, Bosch-Navarrete, C, Musso-Buendia, A, Recio, E, Kaiser, M, Whittingham, J.L, Roberts, S.M, Shevtsov, M, Brannigan, J.A, Kahnberg, P, Brun, R, Wilson, K.S, Gonzalez-Pacanowska, D, Johansson, N.G, Gilbert, I.H. | | Deposit date: | 2011-07-28 | | Release date: | 2012-08-29 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.396 Å) | | Cite: | Design, synthesis, and evaluation of 5'-diphenyl nucleoside analogues as inhibitors of the Plasmodium falciparum dUTPase.
Chemmedchem, 6, 2011
|
|
3T64
 
 | | 5'-Diphenyl Nucleoside Inhibitors of Plasmodium falciparum dUTPase | | Descriptor: | 2',5'-dideoxy-5'-[(diphenylmethyl)amino]uridine, 5'-(BENZHYDRYLAMINO)-2',5'-DIDEOXYURIDINE, Deoxyuridine 5'-triphosphate nucleotidohydrolase, ... | | Authors: | Hampton, S.E, Baragana, B, Schipani, A, Bosch-Navarrete, C, Musso-Buendia, A, Recio, E, Kaiser, M, Whittingham, J.L, Roberts, S.M, Shevtsov, M, Brannigan, J.A, Kahnberg, P, Brun, R, Wilson, K.S, Gonzalez-Pacanowska, D, Johansson, N.G, Gilbert, I.H. | | Deposit date: | 2011-07-28 | | Release date: | 2012-08-29 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Design, synthesis, and evaluation of 5'-diphenyl nucleoside analogues as inhibitors of the Plasmodium falciparum dUTPase.
Chemmedchem, 6, 2011
|
|
3T70
 
 | | 5'-Diphenyl Nucleoside Inhibitors of Plasmodium falciparum dUTPase | | Descriptor: | 2',5'-dideoxy-5'-[(diphenylmethyl)(methyl)amino]uridine, Deoxyuridine 5'-triphosphate nucleotidohydrolase, putative, ... | | Authors: | Hampton, S.E, Baragana, B, Schipani, A, Bosch-Navarrete, C, Musso-Buendia, A, Recio, E, Kaiser, M, Whittingham, J.L, Roberts, S.M, Shevtsov, M, Brannigan, J.A, Kahnberg, P, Brun, R, Wilson, K.S, Gonzalez-Pacanowska, D, Johansson, N.G, Gilbert, I.H. | | Deposit date: | 2011-07-29 | | Release date: | 2012-08-29 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Design, synthesis, and evaluation of 5'-diphenyl nucleoside analogues as inhibitors of the Plasmodium falciparum dUTPase.
Chemmedchem, 6, 2011
|
|
2LZH
 
 | | THE STRUCTURES OF THE MONOCLINIC AND ORTHORHOMBIC FORMS OF HEN EGG-WHITE LYSOZYME AT 6 ANGSTROMS RESOLUTION. | | Descriptor: | HEN EGG WHITE LYSOZYME | | Authors: | Artymiuk, P.J, Blake, C.C.F, Rice, D.W, Wilson, K.S. | | Deposit date: | 1981-06-29 | | Release date: | 1981-09-28 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (6 Å) | | Cite: | The Structures of the Monoclinic and Orthorhombic Forms of Hen Egg-White Lysozyme at 6 Angstroms Resolution.
Acta Crystallogr.,Sect.B, 38, 1982
|
|
1BLU
 
 | | STRUCTURE OF THE 2[4FE-4S] FERREDOXIN FROM CHROMATIUM VINOSUM | | Descriptor: | FERREDOXIN, IRON/SULFUR CLUSTER | | Authors: | Dauter, Z, Wilson, K.S, Sieker, L.C, Moulis, J.M. | | Deposit date: | 1996-04-16 | | Release date: | 1996-11-08 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystal structure of the 2[4Fe-4S] ferredoxin from Chromatium vinosum: evolutionary and mechanistic inferences for [3/4Fe-4S] ferredoxins.
Protein Sci., 5, 1996
|
|
1AY7
 
 | | RIBONUCLEASE SA COMPLEX WITH BARSTAR | | Descriptor: | BARSTAR, GUANYL-SPECIFIC RIBONUCLEASE SA | | Authors: | Sevcik, J, Urbanikova, L, Dauter, Z, Wilson, K.S. | | Deposit date: | 1997-11-14 | | Release date: | 1999-03-02 | | Last modified: | 2023-08-02 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Recognition of RNase Sa by the inhibitor barstar: structure of the complex at 1.7 A resolution.
Acta Crystallogr.,Sect.D, 54, 1998
|
|
1AYX
 
 | | CRYSTAL STRUCTURE OF GLUCOAMYLASE FROM SACCHAROMYCOPSIS FIBULIGERA AT 1.7 ANGSTROMS | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, GLUCOAMYLASE | | Authors: | Sevcik, J, Hostinova, E, Gasperik, J, Solovicova, A, Wilson, K.S, Dauter, Z. | | Deposit date: | 1997-11-12 | | Release date: | 1998-05-13 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structure of glucoamylase from Saccharomycopsis fibuligera at 1.7 A resolution.
Acta Crystallogr.,Sect.D, 54, 1998
|
|
1BOX
 
 | | N39S MUTANT OF RNASE SA FROM STREPTOMYCES AUREOFACIENS | | Descriptor: | GUANYL-SPECIFIC RIBONUCLEASE SA | | Authors: | Hebert, E.J, Giletto, A, Sevcik, J, Urbanikova, L, Wilson, K.S, Dauter, Z, Pace, C.N. | | Deposit date: | 1998-08-07 | | Release date: | 1999-12-29 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Contribution of a conserved asparagine to the conformational stability of ribonucleases Sa, Ba, and T1.
Biochemistry, 37, 1998
|
|
