1SXX
 
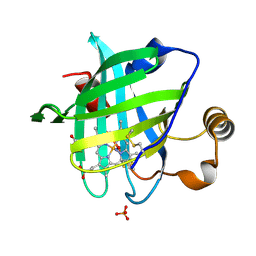 | | 1.0 A Crystal Structure of D129A/L130A Mutant of Nitrophorin 4 Complexed with Nitric Oxide | | Descriptor: | NITRIC OXIDE, Nitrophorin 4, PHOSPHATE ION, ... | | Authors: | Maes, E.M, Weichsel, A, Andersen, J.F, Shepley, D, Montfort, W.R. | | Deposit date: | 2004-03-31 | | Release date: | 2004-06-08 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.01 Å) | | Cite: | Role of binding site loops in controlling nitric oxide release: structure and kinetics of mutant forms of nitrophorin 4
Biochemistry, 43, 2004
|
|
1CLW
 
 | | TAILSPIKE PROTEIN FROM PHAGE P22, V331A MUTANT | | Descriptor: | TAILSPIKE PROTEIN | | Authors: | Steinbacher, S, Baxa, U, Weintraub, A, Huber, R, Seckler, R. | | Deposit date: | 1999-05-04 | | Release date: | 1999-11-05 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Mutations improving the folding of phage P22 tailspike protein affect its receptor binding activity.
J.Mol.Biol., 293, 1999
|
|
6RI7
 
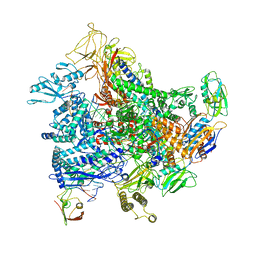 | | Cryo-EM structure of E. coli RNA polymerase elongation complex bound to GreB transcription factor | | Descriptor: | DNA-directed RNA polymerase subunit alpha, DNA-directed RNA polymerase subunit beta, DNA-directed RNA polymerase subunit beta', ... | | Authors: | Abdelkareem, M, Saint-Andre, C, Takacs, M, Papai, G, Crucifix, C, Guo, X, Ortiz, J, Weixlbaumer, A. | | Deposit date: | 2019-04-23 | | Release date: | 2019-07-03 | | Last modified: | 2024-05-22 | | Method: | ELECTRON MICROSCOPY (3.9 Å) | | Cite: | Structural Basis of Transcription: RNA Polymerase Backtracking and Its Reactivation.
Mol.Cell, 75, 2019
|
|
6RH3
 
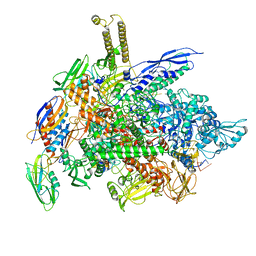 | | Cryo-EM structure of E. coli RNA polymerase elongation complex bound to CTP substrate | | Descriptor: | CYTIDINE-5'-TRIPHOSPHATE, DNA-directed RNA polymerase subunit alpha, DNA-directed RNA polymerase subunit beta, ... | | Authors: | Abdelkareem, M, Saint-Andre, C, Takacs, M, Papai, G, Crucifix, C, Guo, X, Ortiz, J, Weixlbaumer, A. | | Deposit date: | 2019-04-18 | | Release date: | 2019-07-03 | | Last modified: | 2024-05-22 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | Structural Basis of Transcription: RNA Polymerase Backtracking and Its Reactivation.
Mol.Cell, 75, 2019
|
|
6RI9
 
 | | Cryo-EM structure of E. coli RNA polymerase backtracked elongation complex in non-swiveled state | | Descriptor: | DNA-directed RNA polymerase subunit alpha, DNA-directed RNA polymerase subunit beta, DNA-directed RNA polymerase subunit beta', ... | | Authors: | Abdelkareem, M, Saint-Andre, C, Takacs, M, Papai, G, Crucifix, C, Guo, X, Ortiz, J, Weixlbaumer, A. | | Deposit date: | 2019-04-23 | | Release date: | 2019-07-03 | | Last modified: | 2024-05-22 | | Method: | ELECTRON MICROSCOPY (3.7 Å) | | Cite: | Structural Basis of Transcription: RNA Polymerase Backtracking and Its Reactivation.
Mol.Cell, 75, 2019
|
|
6RIN
 
 | | Cryo-EM structure of E. coli RNA polymerase backtracked elongation complex bound to GreB transcription factor | | Descriptor: | DNA-directed RNA polymerase subunit alpha, DNA-directed RNA polymerase subunit beta, DNA-directed RNA polymerase subunit beta', ... | | Authors: | Abdelkareem, M, Saint-Andre, C, Takacs, M, Papai, G, Crucifix, C, Guo, X, Ortiz, J, Weixlbaumer, A. | | Deposit date: | 2019-04-24 | | Release date: | 2019-07-03 | | Last modified: | 2024-05-22 | | Method: | ELECTRON MICROSCOPY (3.7 Å) | | Cite: | Structural Basis of Transcription: RNA Polymerase Backtracking and Its Reactivation.
Mol.Cell, 75, 2019
|
|
1IKE
 
 | | Crystal Structure of Nitrophorin 4 from Rhodnius Prolixus Complexed with Histamine at 1.5 A Resolution | | Descriptor: | HISTAMINE, Nitrophorin 4, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Roberts, S.A, Weichsel, A, Qiu, Y, Shelnutt, J.A, Walker, F.A, Montfort, W.R. | | Deposit date: | 2001-05-03 | | Release date: | 2001-10-03 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Ligand-induced heme ruffling and bent no geometry in ultra-high-resolution structures of nitrophorin 4.
Biochemistry, 40, 2001
|
|
6RIP
 
 | | Cryo-EM structure of E. coli RNA polymerase backtracked elongation complex in swiveled state | | Descriptor: | DNA-directed RNA polymerase subunit alpha, DNA-directed RNA polymerase subunit beta, DNA-directed RNA polymerase subunit beta', ... | | Authors: | Abdelkareem, M, Saint-Andre, C, Takacs, M, Papai, G, Crucifix, C, Guo, X, Ortiz, J, Weixlbaumer, A. | | Deposit date: | 2019-04-24 | | Release date: | 2019-07-03 | | Last modified: | 2024-05-22 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Structural Basis of Transcription: RNA Polymerase Backtracking and Its Reactivation.
Mol.Cell, 75, 2019
|
|
1IKJ
 
 | | 1.27 A CRYSTAL STRUCTURE OF NITROPHORIN 4 FROM RHODNIUS PROLIXUS COMPLEXED WITH IMIDAZOLE | | Descriptor: | IMIDAZOLE, NITROPHORIN 4, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Roberts, S.A, Weichsel, A, Qui, Y, Shelnutt, J.A, Walker, F.A, Montfort, W.R. | | Deposit date: | 2001-05-03 | | Release date: | 2001-10-03 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.27 Å) | | Cite: | Ligand-induced heme ruffling and bent no geometry in ultra-high-resolution structures of nitrophorin 4.
Biochemistry, 40, 2001
|
|
4CR2
 
 | | Deep classification of a large cryo-EM dataset defines the conformational landscape of the 26S proteasome | | Descriptor: | 26S PROTEASE REGULATORY SUBUNIT 4 HOMOLOG, 26S PROTEASE REGULATORY SUBUNIT 6A, 26S PROTEASE REGULATORY SUBUNIT 6B HOMOLOG, ... | | Authors: | Unverdorben, P, Beck, F, Sledz, P, Schweitzer, A, Pfeifer, G, Plitzko, J.M, Baumeister, W, Foerster, F. | | Deposit date: | 2014-02-25 | | Release date: | 2014-04-02 | | Last modified: | 2018-10-03 | | Method: | ELECTRON MICROSCOPY (7.7 Å) | | Cite: | Deep Classification of a Large Cryo-Em Dataset Defines the Conformational Landscape of the 26S Proteasome.
Proc.Natl.Acad.Sci.USA, 111, 2014
|
|
4CR4
 
 | | Deep classification of a large cryo-EM dataset defines the conformational landscape of the 26S proteasome | | Descriptor: | 26S PROTEASE REGULATORY SUBUNIT 4 HOMOLOG, 26S PROTEASE REGULATORY SUBUNIT 6A, 26S PROTEASE REGULATORY SUBUNIT 6B HOMOLOG, ... | | Authors: | Unverdorben, P, Beck, F, Sledz, P, Schweitzer, A, Pfeifer, G, Plitzko, J.M, Baumeister, W, Foerster, F. | | Deposit date: | 2014-02-25 | | Release date: | 2014-04-02 | | Last modified: | 2024-05-08 | | Method: | ELECTRON MICROSCOPY (8.8 Å) | | Cite: | Deep Classification of a Large Cryo-Em Dataset Defines the Conformational Landscape of the 26S Proteasome.
Proc.Natl.Acad.Sci.USA, 111, 2014
|
|
4CR3
 
 | | Deep classification of a large cryo-EM dataset defines the conformational landscape of the 26S proteasome | | Descriptor: | 26S PROTEASE REGULATORY SUBUNIT 4 HOMOLOG, 26S PROTEASE REGULATORY SUBUNIT 6A, 26S PROTEASE REGULATORY SUBUNIT 6B HOMOLOG, ... | | Authors: | Unverdorben, P, Beck, F, Sledz, P, Schweitzer, A, Pfeifer, G, Plitzko, J.M, Baumeister, W, Foerster, F. | | Deposit date: | 2014-02-25 | | Release date: | 2014-04-02 | | Last modified: | 2024-05-08 | | Method: | ELECTRON MICROSCOPY (9.3 Å) | | Cite: | Deep Classification of a Large Cryo-Em Dataset Defines the Conformational Landscape of the 26S Proteasome.
Proc.Natl.Acad.Sci.USA, 111, 2014
|
|
1YWB
 
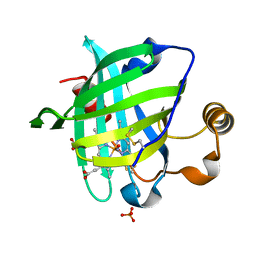 | | 0.9 A Structure of NP4 from Rhodnius Prolixus complexed with NO at pH 5.6 | | Descriptor: | NITRIC OXIDE, PHOSPHATE ION, PROTOPORPHYRIN IX CONTAINING FE, ... | | Authors: | Maes, E.M, Weichsel, A, Roberts, S.A, Montfort, W.R. | | Deposit date: | 2005-02-17 | | Release date: | 2005-10-04 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (0.97 Å) | | Cite: | Ultrahigh Resolution Structures of Nitrophorin 4: Heme Distortion in Ferrous CO and NO Complexes
Biochemistry, 44, 2005
|
|
1AIU
 
 | | HUMAN THIOREDOXIN (D60N MUTANT, REDUCED FORM) | | Descriptor: | THIOREDOXIN | | Authors: | Andersen, J.F, Gasdaska, J.R, Sanders, D.A.R, Weichsel, A, Powis, G, Montfort, W.R. | | Deposit date: | 1997-04-25 | | Release date: | 1997-07-07 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Human thioredoxin homodimers: regulation by pH, role of aspartate 60, and crystal structure of the aspartate 60 --> asparagine mutant.
Biochemistry, 36, 1997
|
|
1YWC
 
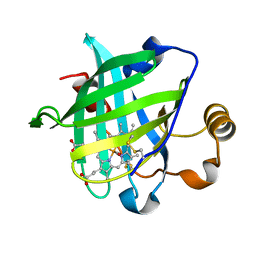 | | Structure of the ferrous CO complex of NP4 from Rhodnius Prolixus at pH 7.0 | | Descriptor: | CARBON MONOXIDE, PROTOPORPHYRIN IX CONTAINING FE, nitrophorin 4 | | Authors: | Maes, E.M, Weichsel, A, Roberts, S.A, Montfort, W.R. | | Deposit date: | 2005-02-17 | | Release date: | 2005-10-04 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1 Å) | | Cite: | Ultrahigh Resolution Structures of Nitrophorin 4: Heme Distortion in Ferrous CO and NO Complexes
Biochemistry, 44, 2005
|
|
1YWD
 
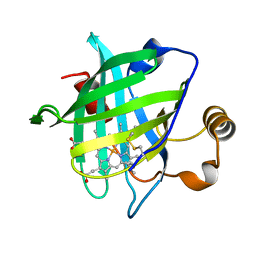 | | 1.08 A Structure of Ferrous NP4 (aquo complex) | | Descriptor: | PROTOPORPHYRIN IX CONTAINING FE, nitrophorin 4 | | Authors: | Maes, E.M, Weichsel, A, Roberts, S.A, Montfort, W.R. | | Deposit date: | 2005-02-17 | | Release date: | 2005-10-04 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.08 Å) | | Cite: | Ultrahigh Resolution Structures of Nitrophorin 4: Heme Distortion in Ferrous CO and NO Complexes
Biochemistry, 44, 2005
|
|
1YWA
 
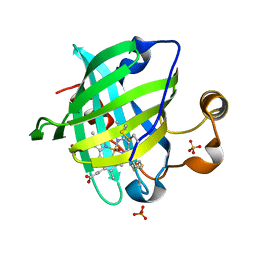 | | 0.9 A Structure of NP4 from Rhodnius Prolixus complexed with CO at pH 5.6 | | Descriptor: | CARBON MONOXIDE, PHOSPHATE ION, PROTOPORPHYRIN IX CONTAINING FE, ... | | Authors: | Maes, E.M, Weichsel, A, Roberts, S.A, Montfort, W.R. | | Deposit date: | 2005-02-17 | | Release date: | 2005-10-04 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (0.89 Å) | | Cite: | Ultrahigh Resolution Structures of Nitrophorin 4: Heme Distortion in Ferrous CO and NO Complexes
Biochemistry, 44, 2005
|
|
3SKX
 
 | | Crystal structure of the ATP binding domain of Archaeoglobus fulgidus COPB | | Descriptor: | ACETATE ION, Copper-exporting P-type ATPase B | | Authors: | Jayakanthan, S, Roberts, S.A, Weichsel, A, Arguello, J.M, McEvoy, M.M. | | Deposit date: | 2011-06-23 | | Release date: | 2012-06-20 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.59 Å) | | Cite: | Conformations of the apo-, substrate-bound and phosphate-bound ATP-binding domain of the Cu(II) ATPase CopB illustrate coupling of domain movement to the catalytic cycle.
Biosci.Rep., 32, 2012
|
|
3SKY
 
 | | 2.1A crystal structure of the phosphate bound ATP binding domain of Archaeoglobus fulgidus COPB | | Descriptor: | 3[N-MORPHOLINO]PROPANE SULFONIC ACID, Copper-exporting P-type ATPase B, PHOSPHATE ION | | Authors: | Jayakanthan, S, Roberts, S.A, Weichsel, A, Arguello, J.M, McEvoy, M.M. | | Deposit date: | 2011-06-23 | | Release date: | 2012-06-20 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Conformations of the apo-, substrate-bound and phosphate-bound ATP-binding domain of the Cu(II) ATPase CopB illustrate coupling of domain movement to the catalytic cycle.
Biosci.Rep., 32, 2012
|
|
3FLL
 
 | | Crystal structure of E55Q mutant of nitrophorin 4 | | Descriptor: | AMMONIA, Nitrophorin-4, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Montfort, W.R, Weichsel, A. | | Deposit date: | 2008-12-18 | | Release date: | 2009-02-10 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Effect of mutation of carboxyl side-chain amino acids near the heme on the midpoint potentials and ligand binding constants of nitrophorin 2 and its NO, histamine, and imidazole complexes.
J.Am.Chem.Soc., 131, 2009
|
|
3EYA
 
 | | Structural basis for membrane binding and catalytic activation of the peripheral membrane enzyme pyruvate oxidase from Escherichia coli | | Descriptor: | FLAVIN-ADENINE DINUCLEOTIDE, MAGNESIUM ION, PHOSPHATE ION, ... | | Authors: | Neumann, P, Weidner, A, Pech, A, Stubbs, M.T, Tittmann, K. | | Deposit date: | 2008-10-20 | | Release date: | 2008-11-04 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structural basis for membrane binding and catalytic activation of the peripheral membrane enzyme pyruvate oxidase from Escherichia coli.
Proc.Natl.Acad.Sci.USA, 105, 2008
|
|
3EY9
 
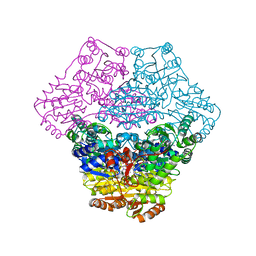 | | Structural basis for membrane binding and catalytic activation of the peripheral membrane enzyme pyruvate oxidase from Escherichia coli | | Descriptor: | FLAVIN-ADENINE DINUCLEOTIDE, MAGNESIUM ION, Pyruvate dehydrogenase [cytochrome], ... | | Authors: | Neumann, P, Weidner, A, Pech, A, Stubbs, M.T, Tittmann, K. | | Deposit date: | 2008-10-20 | | Release date: | 2008-11-04 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Structural basis for membrane binding and catalytic activation of the peripheral membrane enzyme pyruvate oxidase from Escherichia coli.
Proc.Natl.Acad.Sci.USA, 105, 2008
|
|
3FGC
 
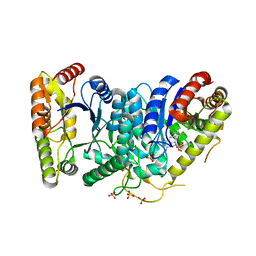 | | Crystal Structure of the Bacterial Luciferase:Flavin Complex Reveals the Basis of Intersubunit Communication | | Descriptor: | Alkanal monooxygenase alpha chain, Alkanal monooxygenase beta chain, FLAVIN MONONUCLEOTIDE, ... | | Authors: | Campbell, Z.T, Weichsel, A, Montfort, W.R, Baldwin, T.O. | | Deposit date: | 2008-12-05 | | Release date: | 2009-05-26 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal structure of the bacterial luciferase/flavin complex provides insight into the function of the beta subunit.
Biochemistry, 48, 2009
|
|
2Y9W
 
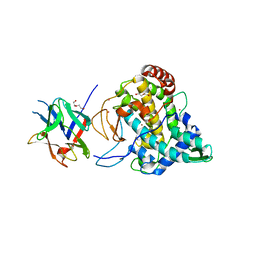 | | Crystal structure of PPO3, a tyrosinase from Agaricus bisporus, in deoxy-form that contains additional unknown lectin-like subunit | | Descriptor: | COPPER (II) ION, DI(HYDROXYETHYL)ETHER, HOLMIUM ATOM, ... | | Authors: | Ismaya, W.T, Rozeboom, H.J, Weijn, A, Mes, J.J, Fusetti, F, Wichers, H.J, Dijkstra, B.W. | | Deposit date: | 2011-02-17 | | Release date: | 2011-07-06 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal Structure of Agaricus Bisporus Mushroom Tyrosinase: Identity of the Tetramer Subunits and Interaction with Tropolone.
Biochemistry, 50, 2011
|
|
2Y9X
 
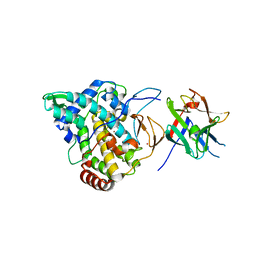 | | Crystal structure of PPO3, a tyrosinase from Agaricus bisporus, in deoxy-form that contains additional unknown lectin-like subunit, with inhibitor tropolone | | Descriptor: | 2-HYDROXYCYCLOHEPTA-2,4,6-TRIEN-1-ONE, COPPER (II) ION, HOLMIUM ATOM, ... | | Authors: | Ismaya, W.T, Rozeboom, H.J, Weijn, A, Mes, J.J, Fusetti, F, Wichers, H.J, Dijkstra, B.W. | | Deposit date: | 2011-02-17 | | Release date: | 2011-07-06 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.78 Å) | | Cite: | Crystal Structure of Agaricus Bisporus Mushroom Tyrosinase: Identity of the Tetramer Subunits and Interaction with Tropolone.
Biochemistry, 50, 2011
|
|
