1HEK
 
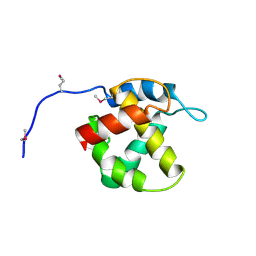 | | Crystal structure of equine infectious anaemia virus matrix antigen (EIAV MA) | | Descriptor: | GAG POLYPROTEIN, CORE PROTEIN P15 | | Authors: | Hatanaka, H, Iourin, O, Rao, Z, Fry, E, Kingsman, A, Stuart, D.I. | | Deposit date: | 2000-11-24 | | Release date: | 2001-11-23 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structure of Equine Infectious Anemia Virus Matrix Protein.
J.Virol., 76, 2002
|
|
8C3V
 
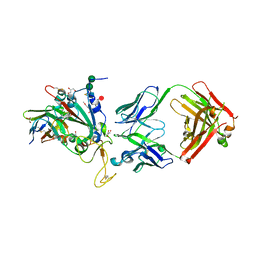 | | SARS-CoV-2 Delta-RBD complexed with BA.2-13 Fab and C1 nanobody | | Descriptor: | 2-(2-METHOXYETHOXY)ETHANOL, BA.2-13 heavy chain, BA.2-13 light chain, ... | | Authors: | Zhou, D, Ren, J, Stuart, D.I. | | Deposit date: | 2022-12-28 | | Release date: | 2023-03-22 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.74 Å) | | Cite: | Rapid escape of new SARS-CoV-2 Omicron variants from BA.2-directed antibody responses.
Cell Rep, 42, 2023
|
|
8CBD
 
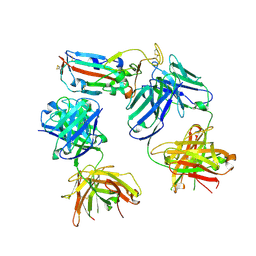 | | SARS-CoV-2 Delta-RBD complexed with BA.4/5-1 and EY6A Fabs | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, BA.4/5-1 heavy chain, BA.4/5-1 light chain, ... | | Authors: | Zhou, D, Ren, J, Stuart, D.I. | | Deposit date: | 2023-01-25 | | Release date: | 2024-02-07 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (3.52 Å) | | Cite: | Emerging variants develop total escape from potent monoclonal antibodies induced by BA.4/5 infection.
Nat Commun, 15, 2024
|
|
8CBF
 
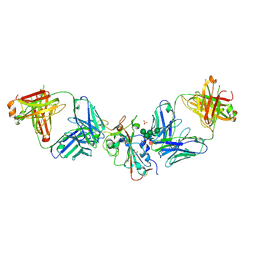 | | SARS-CoV-2 Delta-RBD complexed with Omi-42 and Beta-49 Fabs | | Descriptor: | Beta-49 heavy chain, Beta-49 light chain, CHLORIDE ION, ... | | Authors: | Zhou, D, Ren, J, Stuart, D.I. | | Deposit date: | 2023-01-25 | | Release date: | 2024-02-07 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.33 Å) | | Cite: | Emerging variants develop total escape from potent monoclonal antibodies induced by BA.4/5 infection.
Nat Commun, 15, 2024
|
|
8CBE
 
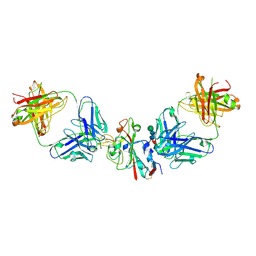 | | SARS-CoV-2 Delta-RBD complexed with BA.4/5-2 and Beta-49 Fabs | | Descriptor: | BA.4/5-2 heavy chain, BA.4/5-2 light chain, Beta-49 heavy chain, ... | | Authors: | Zhou, D, Ren, J, Stuart, D.I. | | Deposit date: | 2023-01-25 | | Release date: | 2024-02-07 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (3.16 Å) | | Cite: | Emerging variants develop total escape from potent monoclonal antibodies induced by BA.4/5 infection.
Nat Commun, 15, 2024
|
|
1ZBE
 
 | | Foot-and Mouth Disease Virus Serotype A1061 | | Descriptor: | Coat protein VP1, Coat protein VP2, Coat protein VP3, ... | | Authors: | Fry, E.E, Newman, J.W, Curry, S, Najjam, S, Jackson, T, Blakemore, W, Lea, S.M, Miller, L, Burman, A, King, A.M, Stuart, D.I. | | Deposit date: | 2005-04-08 | | Release date: | 2005-06-28 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Structure of Foot-and-mouth disease virus serotype A1061 alone and complexed with oligosaccharide receptor: receptor conservation in the face of antigenic variation.
J.Gen.Virol., 86, 2005
|
|
1H15
 
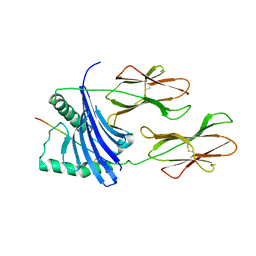 | | X-ray crystal structure of HLA-DRA1*0101/DRB5*0101 complexed with a peptide from Epstein Barr Virus DNA polymerase | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, DNA POLYMERASE, ... | | Authors: | Lang, H, Jacobsen, H, Ikemizu, S, Andersson, C, Harlos, K, Madsen, L, Hjorth, P, Sondergaard, L, Svejgaard, A, Wucherpfennig, K, Stuart, D.I, Bell, J.I, Jones, E.Y, Fugger, L. | | Deposit date: | 2002-07-02 | | Release date: | 2002-10-03 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | A Functional and Structural Basis for Tcr Cross-Reactivity in Multiple Sclerosis
Nat.Immunol., 3, 2002
|
|
2BTV
 
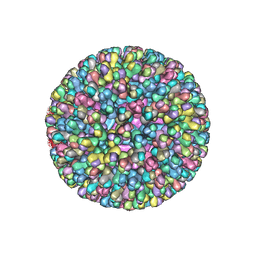 | | ATOMIC MODEL FOR BLUETONGUE VIRUS (BTV) CORE | | Descriptor: | PROTEIN (VP3 CORE PROTEIN), PROTEIN (VP7 CORE PROTEIN) | | Authors: | Grimes, J.M, Burroughs, J.N, Gouet, P, Diprose, J.M, Malby, R, Zientras, S, Mertens, P.P.C, Stuart, D.I. | | Deposit date: | 1998-09-05 | | Release date: | 1998-09-30 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (3.5 Å) | | Cite: | The atomic structure of the bluetongue virus core.
Nature, 395, 1998
|
|
1ZBA
 
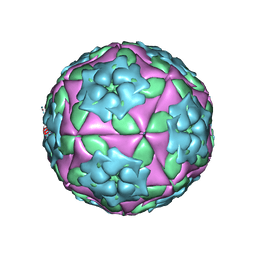 | | Foot-and-Mouth Disease virus serotype A1061 complexed with oligosaccharide receptor. | | Descriptor: | 2-deoxy-6-O-sulfo-2-(sulfoamino)-alpha-D-glucopyranose-(1-4)-2-O-sulfo-alpha-L-idopyranuronic acid-(1-4)-2-deoxy-6-O-sulfo-2-(sulfoamino)-alpha-D-glucopyranose, Coat protein VP1, Coat protein VP2, ... | | Authors: | Fry, E.E, Newman, J.W, Curry, S, Najjam, S, Jackson, T, Blakemore, W, Lea, S.M, Miller, L, Burman, A, King, A.M, Stuart, D.I. | | Deposit date: | 2005-04-08 | | Release date: | 2005-06-28 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structure of Foot-and-mouth disease virus serotype A1061 alone and complexed with oligosaccharide receptor: receptor conservation in the face of antigenic variation.
J.Gen.Virol., 86, 2005
|
|
2BRY
 
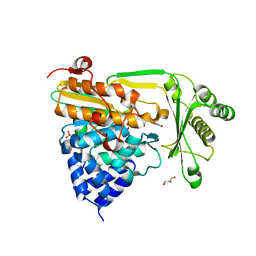 | | Crystal structure of the native monooxygenase domain of MICAL at 1.45 A resolution | | Descriptor: | CHLORIDE ION, FLAVIN-ADENINE DINUCLEOTIDE, GLYCEROL, ... | | Authors: | Siebold, C, Berrow, N, Walter, T.S, Harlos, K, Owens, R.J, Terman, J.R, Stuart, D.I, Kolodkin, A.L, Pasterkamp, R.J, Jones, E.Y. | | Deposit date: | 2005-05-13 | | Release date: | 2005-10-26 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | High-Resolution Structure of the Catalytic Region of Mical (Molecule Interacting with Casl), a Multidomain Flavoenzyme-Signaling Molecule.
Proc.Natl.Acad.Sci.USA, 102, 2005
|
|
2C4C
 
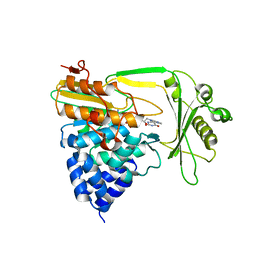 | | Crystal structure of the NADPH-treated monooxygenase domain of MICAL | | Descriptor: | CHLORIDE ION, FLAVIN-ADENINE DINUCLEOTIDE, NEDD9-INTERACTING PROTEIN WITH CALPONIN HOMOLOGY AND LIM DOMAINS | | Authors: | Siebold, C, Berrow, N, Walter, T.S, Harlos, K, Owens, R.J, Terman, J.R, Stuart, D.I, Kolodkin, A.L, Pasterkamp, R.J, Jones, E.Y. | | Deposit date: | 2005-10-18 | | Release date: | 2005-10-26 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | High-Resolution Structure of the Catalytic Region of Mical (Molecule Interacting with Casl), a Multidomain Flavoenzyme-Signaling Molecule.
Proc.Natl.Acad.Sci.USA, 102, 2005
|
|
1ECW
 
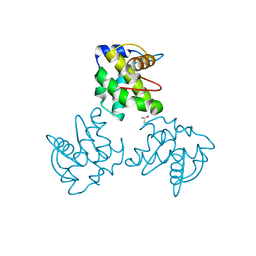 | | CRYSTAL STRUCTURE OF SIMIAN IMMUNODEFICIENCY VIRUS MATRIX ANTIGEN (SIV MA) AT 293K. | | Descriptor: | GAG POLYPROTEIN, ISOPROPYL ALCOHOL | | Authors: | Rao, Z, Belyaev, A, Fry, E, Roy, P, Jones, I.M, Stuart, D.I. | | Deposit date: | 2000-01-26 | | Release date: | 2000-02-16 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal structure of SIV matrix antigen and implications for virus assembly.
Nature, 378, 1995
|
|
1ED1
 
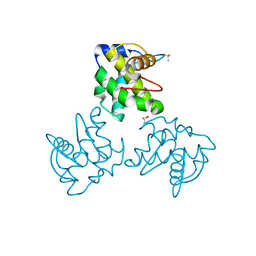 | | CRYSTAL STRUCTURE OF SIMIAN IMMUNODEFICIENCY VIRUS MATRIX ANTIGEN (SIV MA) AT 100K. | | Descriptor: | GAG POLYPROTEIN, ISOPROPYL ALCOHOL | | Authors: | Rao, Z, Belyaev, A, Fry, E, Roy, P, Jones, I.M, Stuart, D.I. | | Deposit date: | 2000-01-26 | | Release date: | 2000-02-16 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystal structure of SIV matrix antigen and implications for virus assembly.
Nature, 378, 1995
|
|
295D
 
 | | CRYSTAL AND SOLUTION STRUCTURES OF THE OLIGONUCLEOTIDE D(ATGCGCAT)2: A COMBINED X-RAY AND NMR STUDY | | Descriptor: | DNA (5'-D(*AP*TP*GP*CP*GP*CP*AP*T)-3') | | Authors: | Clark, G.R, Brown, D.G, Sanderson, M.R, Chwalinski, T, Neidle, S, Veal, J.M, Jones, R.L, Wilson, W.D, Zon, G, Garman, E, Stuart, D.I. | | Deposit date: | 1991-05-28 | | Release date: | 1996-12-04 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Crystal and solution structures of the oligonucleotide d(ATGCGCAT)2: a combined X-ray and NMR study.
Nucleic Acids Res., 18, 1990
|
|
2CDE
 
 | | Structure and binding kinetics of three different human CD1d-alpha- Galactosylceramide specific T cell receptors - iNKT-TCR | | Descriptor: | INKT-TCR | | Authors: | Gadola, S.D, Koch, M, Marles-Wright, J, Lissin, N.M, Sheperd, D, Matulis, G, Harlos, K, Villiger, P.M, Stuart, D.I, Jakobsen, B.K, Cerundolo, V, Jones, E.Y. | | Deposit date: | 2006-01-23 | | Release date: | 2006-03-07 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (3.5 Å) | | Cite: | Structrue and Binding Kinetics of Three Different Human Cd1D-Alpha-Galactosylceramide-Specific T Cell Receptors
J.Exp.Med., 203, 2006
|
|
1RT6
 
 | | HIV-1 REVERSE TRANSCRIPTASE COMPLEXED WITH UC38 | | Descriptor: | 1-METHYL ETHYL 2-CHLORO-5-[[[(1-METHYLETHOXY)THIOOXO]METHYL]AMINO]-BENZOATE, HIV-1 REVERSE TRANSCRIPTASE, PHOSPHATE ION | | Authors: | Ren, J, Stammers, D.K, Stuart, D.I. | | Deposit date: | 1998-07-29 | | Release date: | 1999-07-29 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Crystal structures of HIV-1 reverse transcriptase in complex with carboxanilide derivatives.
Biochemistry, 37, 1998
|
|
1RT7
 
 | | HIV-1 REVERSE TRANSCRIPTASE COMPLEXED WITH UC84 | | Descriptor: | 1-METHYL ETHYL 1-CHLORO-5-[[(5,6DIHYDRO-2-METHYL-1,4-OXATHIIN-3-YL)CARBONYL]AMINO]BENZOATE, HIV-1 REVERSE TRANSCRIPTASE, PHOSPHATE ION | | Authors: | Ren, J, Stammers, D.K, Stuart, D.I. | | Deposit date: | 1998-07-29 | | Release date: | 1999-07-29 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Crystal structures of HIV-1 reverse transcriptase in complex with carboxanilide derivatives.
Biochemistry, 37, 1998
|
|
1RT5
 
 | | HIV-1 REVERSE TRANSCRIPTASE COMPLEXED WITH UC10 | | Descriptor: | HIV-1 REVERSE TRANSCRIPTASE, N-[4-CLORO-3-(T-BUTYLOXOME)PHENYL-2-METHYL-3-FURAN-CARBOTHIAMIDE, PHOSPHATE ION | | Authors: | Ren, J, Stammers, D.K, Stuart, D.I. | | Deposit date: | 1998-07-29 | | Release date: | 1999-07-29 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Crystal structures of HIV-1 reverse transcriptase in complex with carboxanilide derivatives.
Biochemistry, 37, 1998
|
|
1RT4
 
 | | HIV-1 REVERSE TRANSCRIPTASE COMPLEXED WITH UC781 | | Descriptor: | 2-METHYL-FURAN-3-CARBOTHIOIC ACID [4-CHLORO-3-(3-METHYL-BUT-2-ENYLOXY)-PHENYL]-AMIDE, HIV-1 REVERSE TRANSCRIPTASE, PHOSPHATE ION | | Authors: | Ren, J, Stammers, D.K, Stuart, D.I. | | Deposit date: | 1998-07-29 | | Release date: | 1999-07-29 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Crystal structures of HIV-1 reverse transcriptase in complex with carboxanilide derivatives.
Biochemistry, 37, 1998
|
|
3VBS
 
 | | Crystal structure of human Enterovirus 71 | | Descriptor: | Genome Polyprotein, capsid protein VP1, capsid protein VP2, ... | | Authors: | Wang, X, Peng, W, Ren, J, Hu, Z, Xu, J, Lou, Z, Li, X, Yin, W, Shen, X, Porta, C, Walter, T.S, Evans, G, Axford, D, Owen, R, Rowlands, D.J, Wang, J, Stuart, D.I, Fry, E.E, Rao, Z. | | Deposit date: | 2012-01-02 | | Release date: | 2012-02-29 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | A sensor-adaptor mechanism for enterovirus uncoating from structures of EV71.
Nat.Struct.Mol.Biol., 19, 2012
|
|
3VBR
 
 | | Crystal structure of formaldehyde treated empty human Enterovirus 71 particle (room temperature) | | Descriptor: | Genome Polyprotein, capsid protein VP0, capsid protein VP1, ... | | Authors: | Wang, X, Peng, W, Ren, J, Hu, Z, Xu, J, Lou, Z, Li, X, Yin, W, Shen, X, Porta, C, Walter, T.S, Evans, G, Axford, D, Owen, R, Rowlands, D.J, Wang, J, Stuart, D.I, Fry, E.E, Rao, Z. | | Deposit date: | 2012-01-02 | | Release date: | 2012-02-29 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (3.8 Å) | | Cite: | A sensor-adaptor mechanism for enterovirus uncoating from structures of EV71.
Nat.Struct.Mol.Biol., 19, 2012
|
|
3V6F
 
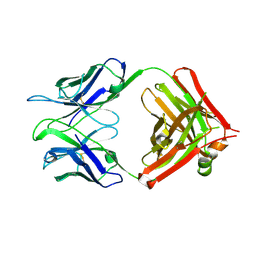 | | Crystal Structure of an anti-HBV e-antigen monoclonal Fab fragment (e6), unbound | | Descriptor: | Fab e6 Heavy Chain, Fab e6 Light Chain | | Authors: | Dimattia, M.A, Watts, N.R, Stahl, S.J, Grimes, J.M, Steven, A.C, Stuart, D.I, Wingfield, P.T. | | Deposit date: | 2011-12-19 | | Release date: | 2013-02-06 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.52 Å) | | Cite: | Antigenic switching of hepatitis B virus by alternative dimerization of the capsid protein.
Structure, 21, 2013
|
|
3VBF
 
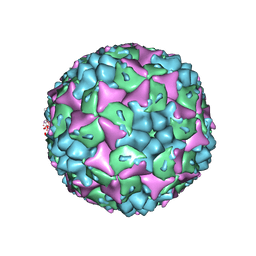 | | Crystal structure of formaldehyde treated human Enterovirus 71 (space group I23) | | Descriptor: | ADENOSINE MONOPHOSPHATE, CHLORIDE ION, Genome Polyprotein, ... | | Authors: | Wang, X, Peng, W, Ren, J, Hu, Z, Xu, J, Lou, Z, Li, X, Yin, W, Shen, X, Porta, C, Walter, T.S, Evans, G, Axford, D, Owen, R, Rowlands, D.J, Wang, J, Stuart, D.I, Fry, E.E, Rao, Z. | | Deposit date: | 2012-01-02 | | Release date: | 2012-02-29 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | A sensor-adaptor mechanism for enterovirus uncoating from structures of EV71.
Nat.Struct.Mol.Biol., 19, 2012
|
|
3VBO
 
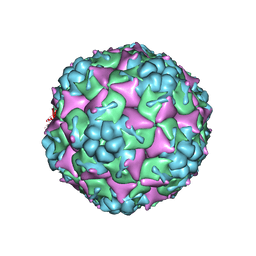 | | Crystal structure of formaldehyde treated empty human Enterovirus 71 particle (cryo at 100K) | | Descriptor: | Genome Polyprotein, capsid protein VP1, capsid protein VP2, ... | | Authors: | Wang, X, Peng, W, Ren, J, Hu, Z, Xu, J, Lou, Z, Li, X, Yin, W, Shen, X, Porta, C, Walter, T.S, Evans, G, Axford, D, Owen, R, Rowlands, D.J, Wang, J, Stuart, D.I, Fry, E.E, Rao, Z. | | Deposit date: | 2012-01-02 | | Release date: | 2012-02-29 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.88 Å) | | Cite: | A sensor-adaptor mechanism for enterovirus uncoating from structures of EV71.
Nat.Struct.Mol.Biol., 19, 2012
|
|
3V6Z
 
 | | Crystal Structure of Hepatitis B Virus e-antigen | | Descriptor: | Fab e6 Heavy Chain, Fab e6 Light Chain, e-antigen | | Authors: | Dimattia, M.A, Watts, N.R, Stahl, S.J, Grimes, J.M, Steven, A.C, Stuart, D.I, Wingfield, P.T. | | Deposit date: | 2011-12-20 | | Release date: | 2013-02-06 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (3.34 Å) | | Cite: | Antigenic switching of hepatitis B virus by alternative dimerization of the capsid protein.
Structure, 21, 2013
|
|
