7W6F
 
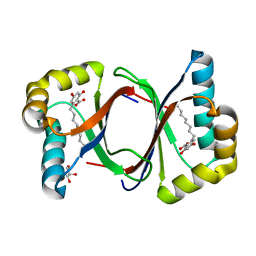 | | Polyketide cyclase OAC-F24I mutant from Cannabis sativa in complex with 6-nonylresorcylic acid | | Descriptor: | 2-nonyl-4,6-bis(oxidanyl)benzoic acid, GLYCEROL, Olivetolic acid cyclase | | Authors: | Nakashima, Y, Lee, Y.E, Morita, H. | | Deposit date: | 2021-12-01 | | Release date: | 2022-02-02 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.58 Å) | | Cite: | Dual Engineering of Olivetolic Acid Cyclase and Tetraketide Synthase to Generate Longer Alkyl-Chain Olivetolic Acid Analogs.
Org.Lett., 24, 2022
|
|
7W6G
 
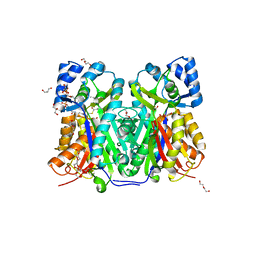 | | TKS-L190G mutant from Cannabis sativa in complex with lauroyl-CoA | | Descriptor: | 3,5,7-trioxododecanoyl-CoA synthase, DI(HYDROXYETHYL)ETHER, GLYCEROL, ... | | Authors: | Nakashima, Y, Lee, Y.E, Morita, H. | | Deposit date: | 2021-12-01 | | Release date: | 2022-02-02 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Dual Engineering of Olivetolic Acid Cyclase and Tetraketide Synthase to Generate Longer Alkyl-Chain Olivetolic Acid Analogs.
Org.Lett., 24, 2022
|
|
7W6E
 
 | |
7W6D
 
 | |
6IGJ
 
 | | Crystal structure of FT condition 4 | | Descriptor: | MAGNESIUM ION, Protein FLOWERING LOCUS T | | Authors: | Watanabe, S, Nakamura, Y, Kanehara, K, Inaba, K. | | Deposit date: | 2018-09-25 | | Release date: | 2019-12-25 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.501 Å) | | Cite: | High-Resolution Crystal Structure of Arabidopsis FLOWERING LOCUS T Illuminates Its Phospholipid-Binding Site in Flowering.
Iscience, 21, 2019
|
|
6IGG
 
 | | Crystal structure of FT condition 1 | | Descriptor: | 1,2-ETHANEDIOL, Protein FLOWERING LOCUS T | | Authors: | Watanabe, S, Nakamura, Y, Kanehara, K, Inaba, K. | | Deposit date: | 2018-09-25 | | Release date: | 2019-12-25 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1 Å) | | Cite: | High-Resolution Crystal Structure of Arabidopsis FLOWERING LOCUS T Illuminates Its Phospholipid-Binding Site in Flowering.
Iscience, 21, 2019
|
|
5ZTB
 
 | | Structure of Sulfurtransferase | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, GLYCEROL, IRON/SULFUR CLUSTER, ... | | Authors: | Tanaka, Y, Chen, M, Narai, S, Yao, M. | | Deposit date: | 2018-05-02 | | Release date: | 2019-06-12 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | The [4Fe-4S] cluster of sulfurtransferase TtuA desulfurizes TtuB during tRNA modification in Thermus thermophilus.
Commun Biol, 3, 2020
|
|
6IGH
 
 | | Crystal structure of FT condition3 | | Descriptor: | 1,2-ETHANEDIOL, Protein FLOWERING LOCUS T | | Authors: | Watanabe, S, Nakamura, Y, Kanehara, K, Inaba, K. | | Deposit date: | 2018-09-25 | | Release date: | 2019-12-25 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.01 Å) | | Cite: | High-Resolution Crystal Structure of Arabidopsis FLOWERING LOCUS T Illuminates Its Phospholipid-Binding Site in Flowering.
Iscience, 21, 2019
|
|
6IGI
 
 | | Crystal structure of FT condition 2 | | Descriptor: | 1,2-ETHANEDIOL, Protein FLOWERING LOCUS T | | Authors: | Watanabe, S, Nakamura, Y, Kanehara, K, Inaba, K. | | Deposit date: | 2018-09-25 | | Release date: | 2019-12-25 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.33 Å) | | Cite: | High-Resolution Crystal Structure of Arabidopsis FLOWERING LOCUS T Illuminates Its Phospholipid-Binding Site in Flowering.
Iscience, 21, 2019
|
|
2ZY6
 
 | | Crystal structure of a truncated tRNA, TPHE39A | | Descriptor: | CALCIUM ION, CHLORIDE ION, MAGNESIUM ION, ... | | Authors: | Tanaka, I, Yao, M, Tanaka, Y, Kitago, Y, Ymagata, S. | | Deposit date: | 2009-01-14 | | Release date: | 2009-06-30 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Deduced RNA binding mechanism of ThiI based on structural and binding analyses of a minimal RNA ligand
Rna, 15, 2009
|
|
3ANZ
 
 | | Crystal Structure of alpha-hemolysin | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, ACETIC ACID, Alpha-hemolysin | | Authors: | Yamashita, K, Kawauchi, H, Tanaka, Y, Yao, M, Tanaka, I. | | Deposit date: | 2010-09-16 | | Release date: | 2011-06-22 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.303 Å) | | Cite: | 2-Methyl-2,4-pentanediol induces spontaneous assembly of staphylococcal alpha-hemolysin into heptameric pore structure
Protein Sci., 20, 2011
|
|
2Z6F
 
 | |
7DYT
 
 | | Human JMJD5 in complex with MN and 5-((4-methoxybenzyl)amino)pyridine-2,4-dicarboxylic acid. | | Descriptor: | 5-((4-methoxybenzyl)amino)pyridine-2,4-dicarboxylic acid, Bifunctional peptidase and arginyl-hydroxylase JMJD5, MANGANESE (II) ION | | Authors: | Nakashima, Y, Brewitz, L, Schofield, C.J. | | Deposit date: | 2021-01-23 | | Release date: | 2022-02-02 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (1.62 Å) | | Cite: | 5-Substituted Pyridine-2,4-dicarboxylate Derivatives Have Potential for Selective Inhibition of Human Jumonji-C Domain-Containing Protein 5.
J.Med.Chem., 66, 2023
|
|
7DYU
 
 | | Human JMJD5 in complex with MN and 5-((4-phenylbutyl)amino)pyridine-2,4-dicarboxylic acid. | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, 5-(4-phenylbutylamino)pyridine-2,4-dicarboxylic acid, Bifunctional peptidase and arginyl-hydroxylase JMJD5, ... | | Authors: | Nakashima, Y, Brewitz, L, Schofield, C.J. | | Deposit date: | 2021-01-23 | | Release date: | 2022-02-02 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (1.72 Å) | | Cite: | 5-Substituted Pyridine-2,4-dicarboxylate Derivatives Have Potential for Selective Inhibition of Human Jumonji-C Domain-Containing Protein 5.
J.Med.Chem., 66, 2023
|
|
7DYX
 
 | | Human JMJD5 in complex with MN and 5-((2-cyclopropylbenzyl)amino)pyridine-2,4-dicarboxylic acid. | | Descriptor: | 5-((2-cyclopropylbenzyl)amino)pyridine-2,4-dicarboxylic acid, Bifunctional peptidase and arginyl-hydroxylase JMJD5, MANGANESE (II) ION | | Authors: | Nakashima, Y, Brewitz, L, Schofield, C.J. | | Deposit date: | 2021-01-23 | | Release date: | 2022-02-02 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (2.27 Å) | | Cite: | 5-Substituted Pyridine-2,4-dicarboxylate Derivatives Have Potential for Selective Inhibition of Human Jumonji-C Domain-Containing Protein 5.
J.Med.Chem., 66, 2023
|
|
7E6J
 
 | | Aspartyl/Asparaginyl beta-hydroxylase (AspH) H725A in complex with Factor X peptide fragment (39mer-4Ser) | | Descriptor: | Aspartyl/asparaginyl beta-hydroxylase, GLYCEROL, PROPANOIC ACID, ... | | Authors: | Nakashima, Y, Brasnett, A, Brewitz, L, Schofield, C.J. | | Deposit date: | 2021-02-22 | | Release date: | 2021-06-30 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Human Oxygenase Variants Employing a Single Protein Fe II Ligand Are Catalytically Active.
Angew.Chem.Int.Ed.Engl., 60, 2021
|
|
7DYV
 
 | | Human JMJD5 in complex with MN and 5-(benzylamino)pyridine-2,4-dicarboxylic acid. | | Descriptor: | 5-(benzylamino)pyridine-2,4-dicarboxylic acid, Bifunctional peptidase and arginyl-hydroxylase JMJD5, MANGANESE (II) ION | | Authors: | Nakashima, Y, Brewitz, L, Schofield, C.J. | | Deposit date: | 2021-01-23 | | Release date: | 2022-02-02 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (1.92 Å) | | Cite: | 5-Substituted Pyridine-2,4-dicarboxylate Derivatives Have Potential for Selective Inhibition of Human Jumonji-C Domain-Containing Protein 5.
J.Med.Chem., 66, 2023
|
|
7DYW
 
 | | Human JMJD5 in complex with MN and 5-((2-methoxybenzyl)amino)pyridine-2,4-dicarboxylic acid. | | Descriptor: | 5-((2-methoxybenzyl)amino)pyridine-2,4-dicarboxylic acid, Bifunctional peptidase and arginyl-hydroxylase JMJD5, MANGANESE (II) ION | | Authors: | Nakashima, Y, Brewitz, L, Schofield, C.J. | | Deposit date: | 2021-01-23 | | Release date: | 2022-02-02 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (2.13 Å) | | Cite: | 5-Substituted Pyridine-2,4-dicarboxylate Derivatives Have Potential for Selective Inhibition of Human Jumonji-C Domain-Containing Protein 5.
J.Med.Chem., 66, 2023
|
|
3A6J
 
 | | E122Q mutant creatininase complexed with creatine | | Descriptor: | Creatinine amidohydrolase, N-[(E)-AMINO(IMINO)METHYL]-N-METHYLGLYCINE, SULFATE ION, ... | | Authors: | Nakajima, Y, Yamashita, K, Ito, K, Yoshimoto, T. | | Deposit date: | 2009-09-02 | | Release date: | 2010-02-09 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Substitution of Glu122 by glutamine revealed the function of the second water molecule as a proton donor in the binuclear metal enzyme creatininase
J.Mol.Biol., 396, 2010
|
|
3A6D
 
 | | Creatininase complexed with 1-methylguanidine | | Descriptor: | 1-METHYLGUANIDINE, Creatinine amidohydrolase, MANGANESE (II) ION, ... | | Authors: | Nakajima, Y, Yamashita, K, Ito, K, Yoshimoto, T. | | Deposit date: | 2009-08-31 | | Release date: | 2010-02-09 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Substitution of Glu122 by glutamine revealed the function of the second water molecule as a proton donor in the binuclear metal enzyme creatininase
J.Mol.Biol., 396, 2010
|
|
3A6E
 
 | | W174F mutant creatininase, type I | | Descriptor: | CACODYLATE ION, Creatinine amidohydrolase, MANGANESE (II) ION, ... | | Authors: | Nakajima, Y, Yamashita, K, Ito, K, Yoshimoto, T. | | Deposit date: | 2009-08-31 | | Release date: | 2010-02-09 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Substitution of Glu122 by glutamine revealed the function of the second water molecule as a proton donor in the binuclear metal enzyme creatininase
J.Mol.Biol., 396, 2010
|
|
3A6L
 
 | | E122Q mutant creatininase, Zn-Zn type | | Descriptor: | CHLORIDE ION, Creatinine amidohydrolase, ZINC ION | | Authors: | Nakajima, Y, Yamashita, K, Ito, K, Yoshimoto, T. | | Deposit date: | 2009-09-02 | | Release date: | 2010-02-09 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Substitution of Glu122 by glutamine revealed the function of the second water molecule as a proton donor in the binuclear metal enzyme creatininase
J.Mol.Biol., 396, 2010
|
|
3A6H
 
 | | W154A mutant creatininase | | Descriptor: | CHLORIDE ION, Creatinine amidohydrolase, MANGANESE (II) ION, ... | | Authors: | Nakajima, Y, Yamashita, K, Ito, K, Yoshimoto, T. | | Deposit date: | 2009-08-31 | | Release date: | 2010-02-09 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Substitution of Glu122 by glutamine revealed the function of the second water molecule as a proton donor in the binuclear metal enzyme creatininase
J.Mol.Biol., 396, 2010
|
|
3A6K
 
 | | The E122Q mutant creatininase, Mn-Zn type | | Descriptor: | CHLORIDE ION, Creatinine amidohydrolase, MANGANESE (II) ION, ... | | Authors: | Nakajima, Y, Yamashita, K, Ito, K, Yoshimoto, T. | | Deposit date: | 2009-09-02 | | Release date: | 2010-02-09 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Substitution of Glu122 by glutamine revealed the function of the second water molecule as a proton donor in the binuclear metal enzyme creatininase
J.Mol.Biol., 396, 2010
|
|
3ANQ
 
 | | human DYRK1A/inhibitor complex | | Descriptor: | (1Z)-1-(3-ethyl-5-hydroxy-1,3-benzothiazol-2(3H)-ylidene)propan-2-one, Dual specificity tyrosine-phosphorylation-regulated kinase 1A | | Authors: | Nonaka, Y, Hosoya, T, Hagiwara, M, Ito, N. | | Deposit date: | 2010-09-06 | | Release date: | 2010-11-10 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Development of a novel selective inhibitor of the Down syndrome-related kinase Dyrk1A
Nat Commun, 1, 2010
|
|
