4TN1
 
 | |
4TMT
 
 | | Translation initiation factor eIF5B (517-858) mutant D533A from C. thermophilum, bound to GTPgammaS | | Descriptor: | 1,2-ETHANEDIOL, 2,3-DIHYDROXY-1,4-DITHIOBUTANE, 5'-GUANOSINE-DIPHOSPHATE-MONOTHIOPHOSPHATE, ... | | Authors: | Kuhle, B, Ficner, R. | | Deposit date: | 2014-06-02 | | Release date: | 2014-09-24 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.58 Å) | | Cite: | A monovalent cation acts as structural and catalytic cofactor in translational GTPases.
Embo J., 33, 2014
|
|
4TMX
 
 | | Translation initiation factor eIF5B (517-858) mutant D533N from C. thermophilum, bound to GTP and sodium | | Descriptor: | 1,2-ETHANEDIOL, ACETIC ACID, GUANOSINE-5'-TRIPHOSPHATE, ... | | Authors: | Kuhle, B, Ficner, R. | | Deposit date: | 2014-06-02 | | Release date: | 2014-09-24 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | A monovalent cation acts as structural and catalytic cofactor in translational GTPases.
Embo J., 33, 2014
|
|
3EWI
 
 | | Structural analysis of the C-terminal domain of murine CMP-Sialic acid Synthetase | | Descriptor: | N-acylneuraminate cytidylyltransferase | | Authors: | Oschlies, M, Dickmanns, A, Stummeyer, K, Gerardy-Schahn, R, Ficner, R, Muenster-Kuehnel, A.K. | | Deposit date: | 2008-10-15 | | Release date: | 2009-08-18 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | A C-terminal phosphatase module conserved in vertebrate CMP-sialic acid synthetases provides a tetramerization interface for the physiologically active enzyme.
J.Mol.Biol., 393, 2009
|
|
4KR7
 
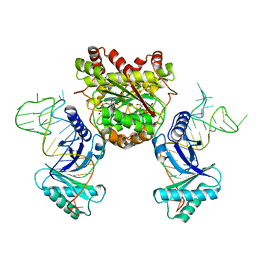 | | Crystal structure of a 4-thiouridine synthetase - RNA complex with bound ATP | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, MAGNESIUM ION, Probable tRNA sulfurtransferase, ... | | Authors: | Neumann, P, Ficner, R, Lakomek, K. | | Deposit date: | 2013-05-16 | | Release date: | 2014-04-23 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (3.421 Å) | | Cite: | Crystal structure of a 4-thiouridine synthetase-RNA complex reveals specificity of tRNA U8 modification.
Nucleic Acids Res., 42, 2014
|
|
4N6Q
 
 | | Crystal structure of VosA velvet domain | | Descriptor: | IODIDE ION, NITRATE ION, VosA | | Authors: | Ahmed, Y.L, Dickmanns, A, Neumann, P, Ficner, R. | | Deposit date: | 2013-10-14 | | Release date: | 2014-01-22 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.79 Å) | | Cite: | The Velvet Family of Fungal Regulators Contains a DNA-Binding Domain Structurally Similar to NF-kappa B.
Plos Biol., 11, 2013
|
|
4N3S
 
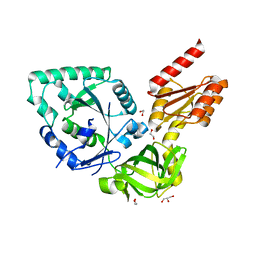 | |
4NWI
 
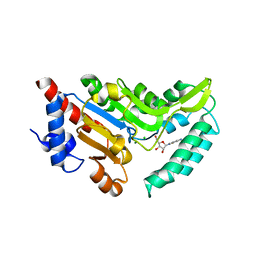 | | Crystal structure of cytosolic 5'-nucleotidase IIIB (cN-IIIB) bound to cytidine | | Descriptor: | 4-AMINO-1-BETA-D-RIBOFURANOSYL-2(1H)-PYRIMIDINONE, 7-methylguanosine phosphate-specific 5'-nucleotidase, CHLORIDE ION, ... | | Authors: | Monecke, T, Neumann, P, Ficner, R. | | Deposit date: | 2013-12-06 | | Release date: | 2014-03-19 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Crystal Structures of the Novel Cytosolic 5'-Nucleotidase IIIB Explain Its Preference for m7GMP
Plos One, 9, 2014
|
|
4N3G
 
 | |
4NCF
 
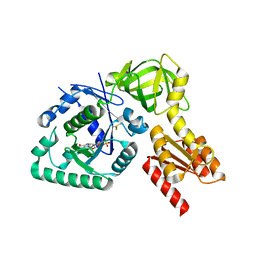 | |
4NOX
 
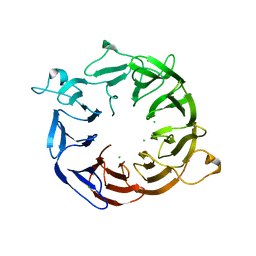 | | Structure of the nine-bladed beta-propeller of eIF3b | | Descriptor: | CHLORIDE ION, Eukaryotic translation initiation factor 3 subunit B | | Authors: | Liu, Y, Neumann, P, Kuhle, B, Monecke, T, Ficner, R. | | Deposit date: | 2013-11-20 | | Release date: | 2014-09-17 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.722 Å) | | Cite: | Translation initiation factor eIF3b contains a nine-bladed beta-propeller and interacts with the 40S ribosomal subunit
Structure, 22, 2014
|
|
4NV0
 
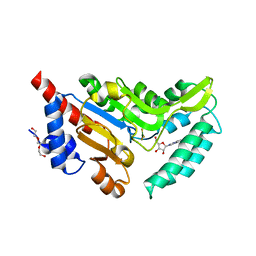 | | Crystal structure of cytosolic 5'-nucleotidase IIIB (cN-IIIB) bound to 7-methylguanosine | | Descriptor: | 7-METHYLGUANOSINE, 7-methylguanosine phosphate-specific 5'-nucleotidase, MAGNESIUM ION, ... | | Authors: | Monecke, T, Neumann, P, Ficner, R. | | Deposit date: | 2013-12-04 | | Release date: | 2014-03-19 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Crystal Structures of the Novel Cytosolic 5'-Nucleotidase IIIB Explain Its Preference for m7GMP
Plos One, 9, 2014
|
|
4N3N
 
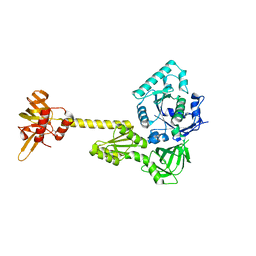 | |
4NCN
 
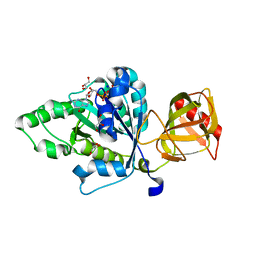 | |
4NCL
 
 | |
3GUD
 
 | | Crystal structure of a novel intramolecular chaperon | | Descriptor: | BROMIDE ION, CHLORIDE ION, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Schulz, E.C, Dickmanns, A, Ficner, R. | | Deposit date: | 2009-03-29 | | Release date: | 2010-02-02 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal structure of an intramolecular chaperone mediating triple-beta-helix folding.
Nat.Struct.Mol.Biol., 17, 2010
|
|
3GVL
 
 | | Crystal Structure of endo-neuraminidaseNF | | Descriptor: | Endo-N-acetylneuraminidase, N-acetyl-alpha-neuraminic acid-(2-8)-N-acetyl-beta-neuraminic acid, N-acetyl-beta-neuraminic acid | | Authors: | Schulz, E.C, Dickmanns, A, Ficner, R. | | Deposit date: | 2009-03-31 | | Release date: | 2010-03-02 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.41 Å) | | Cite: | Structural basis for the recognition and cleavage of polysialic acid by the bacteriophage K1F tailspike protein EndoNF.
J.Mol.Biol., 397, 2010
|
|
3GW6
 
 | | Intramolecular Chaperone | | Descriptor: | BROMIDE ION, CALCIUM ION, CHLORIDE ION, ... | | Authors: | Schulz, E.C, Dickmanns, A, Ficner, R. | | Deposit date: | 2009-03-31 | | Release date: | 2010-02-02 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Crystal structure of an intramolecular chaperone mediating triple-beta-helix folding.
Nat.Struct.Mol.Biol., 17, 2010
|
|
3GVJ
 
 | | Crystal structure of an endo-neuraminidaseNF mutant | | Descriptor: | Endo-N-acetylneuraminidase, N-acetyl-alpha-neuraminic acid-(2-8)-N-acetyl-alpha-neuraminic acid-(2-8)-N-acetyl-alpha-neuraminic acid-(2-8)-N-acetyl-alpha-neuraminic acid-(2-8)-N-acetyl-beta-neuraminic acid, N-acetyl-alpha-neuraminic acid-(2-8)-N-acetyl-alpha-neuraminic acid-(2-8)-N-acetyl-alpha-neuraminic acid-(2-8)-N-acetyl-beta-neuraminic acid | | Authors: | Schulz, E.C, Dickmanns, A, Ficner, R. | | Deposit date: | 2009-03-31 | | Release date: | 2010-03-02 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.48 Å) | | Cite: | Structural basis for the recognition and cleavage of polysialic acid by the bacteriophage K1F tailspike protein EndoNF.
J.Mol.Biol., 397, 2010
|
|
3GVK
 
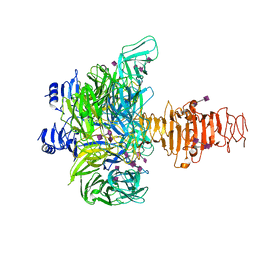 | | Crystal structure of endo-neuraminidase NF mutant | | Descriptor: | Endo-N-acetylneuraminidase, N-acetyl-alpha-neuraminic acid-(2-8)-N-acetyl-alpha-neuraminic acid-(2-8)-N-acetyl-beta-neuraminic acid, N-acetyl-alpha-neuraminic acid-(2-8)-N-acetyl-beta-neuraminic acid, ... | | Authors: | Schulz, E.C, Dickmanns, A, Ficner, R. | | Deposit date: | 2009-03-31 | | Release date: | 2010-03-02 | | Last modified: | 2021-10-13 | | Method: | X-RAY DIFFRACTION (1.84 Å) | | Cite: | Structural basis for the recognition and cleavage of polysialic acid by the bacteriophage K1F tailspike protein EndoNF.
J.Mol.Biol., 397, 2010
|
|
3G38
 
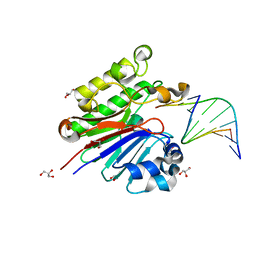 | | The catalytically inactive mutant Mth0212 (D151N) in complex with an 8 bp dsDNA | | Descriptor: | 5'-D(*CP*CP*TP*GP*UP*GP*CP*GP*AP*T)-3', 5'-D(*CP*GP*CP*GP*CP*AP*GP*GP*C)-3', Exodeoxyribonuclease, ... | | Authors: | Lakomek, K, Dickmanns, A, Ficner, R. | | Deposit date: | 2009-02-02 | | Release date: | 2010-03-09 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (3.04 Å) | | Cite: | Crystal Structure Analysis of DNA Uridine Endonuclease Mth212 Bound to DNA
J.Mol.Biol., 399, 2010
|
|
3CTR
 
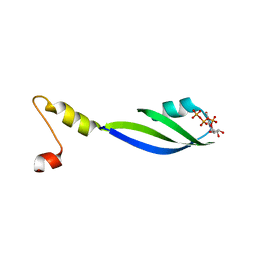 | | Crystal structure of the RRM-domain of the poly(A)-specific ribonuclease PARN bound to m7GTP | | Descriptor: | 7-METHYL-GUANOSINE-5'-TRIPHOSPHATE, Poly(A)-specific ribonuclease PARN | | Authors: | Monecke, T, Schell, S, Dickmanns, A, Ficner, R. | | Deposit date: | 2008-04-14 | | Release date: | 2008-07-29 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystal structure of the RRM domain of poly(A)-specific ribonuclease reveals a novel m(7)G-cap-binding mode.
J.Mol.Biol., 382, 2008
|
|
3G2D
 
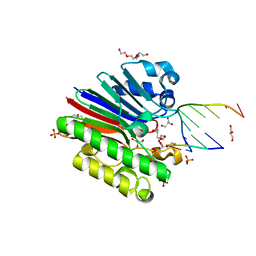 | | Complex of Mth0212 and a 4 bp dsDNA with 3'-overhang | | Descriptor: | 5'-D(*CP*CP*TP*GP*UP*GP*CP*GP*AP*T)-3', 5'-D(*CP*GP*CP*G*CP*AP*GP*GP*C)-3', Exodeoxyribonuclease, ... | | Authors: | Lakomek, K, Dickmanns, A, Ficner, R. | | Deposit date: | 2009-01-31 | | Release date: | 2010-03-09 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal Structure Analysis of DNA Uridine Endonuclease Mth212 Bound to DNA
J.Mol.Biol., 399, 2010
|
|
3G2C
 
 | | Mth0212 in complex with a short ssDNA (CGTA) | | Descriptor: | 5'-D(P*CP*GP*TP*A)-3', Exodeoxyribonuclease, GLYCEROL, ... | | Authors: | Lakomek, K, Dickmanns, A, Ficner, R. | | Deposit date: | 2009-01-31 | | Release date: | 2010-03-09 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal Structure Analysis of DNA Uridine Endonuclease Mth212 Bound to DNA
J.Mol.Biol., 399, 2010
|
|
3G4T
 
 | | Mth0212 (WT) in complex with a 7bp dsDNA | | Descriptor: | 5'-D(*CP*G*TP*AP*CP*TP*AP*CP*G)-3', 5'-D(*CP*GP*TP*AP*(UPS)P*TP*AP*CP*G)-3', Exodeoxyribonuclease, ... | | Authors: | Lakomek, K, Dickmanns, A, Ficner, R. | | Deposit date: | 2009-02-04 | | Release date: | 2010-03-09 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.64 Å) | | Cite: | Crystal Structure Analysis of DNA Uridine Endonuclease Mth212 Bound to DNA
J.Mol.Biol., 399, 2010
|
|
