2YC5
 
 | | Inhibitors of the herbicidal target IspD | | Descriptor: | 2-C-METHYL-D-ERYTHRITOL 4-PHOSPHATE CYTIDYLYLTRANSFERASE, CHLOROPLASTIC, 5-chloro-7-hydroxy-6-(phenylmethyl)pyrazolo[1,5-a]pyrimidine-3-carbonitrile, ... | | Authors: | Hoeffken, H.W. | | Deposit date: | 2011-03-11 | | Release date: | 2011-08-24 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Inhibitors of the Herbicidal Target Ispd: Allosteric Site Binding.
Angew.Chem.Int.Ed.Engl., 50, 2011
|
|
2YC3
 
 | | Inhibitors of the herbicidal target IspD | | Descriptor: | 2-C-METHYL-D-ERYTHRITOL 4-PHOSPHATE CYTIDYLYLTRANSFERASE, CHLOROPLASTIC, 6-benzyl-5-chloro[1,2,4]triazolo[1,5-a]pyrimidin-7(3H)-one, ... | | Authors: | Hoeffken, H.W. | | Deposit date: | 2011-03-11 | | Release date: | 2011-08-24 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Inhibitors of the Herbicidal Target Ispd: Allosteric Site Binding.
Angew.Chem.Int.Ed.Engl., 50, 2011
|
|
2K3I
 
 | | Solution NMR structure of protein yiiS from Shigella flexneri. Northeast Structural Genomics Consortium target SfR90 | | Descriptor: | Uncharacterized protein yiiS | | Authors: | Mills, J.L, Singarapu, K.K, Eletsky, A, Sukumaran, D.K, Wang, D, Jiang, M, Ciccosanti, C, Xiao, R, Liu, J, Baran, M.C, Swapna, G.V.T, Acton, T, Rost, B, Montelione, G.T, Szyperski, T, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2008-05-08 | | Release date: | 2008-07-08 | | Last modified: | 2024-05-08 | | Method: | SOLUTION NMR | | Cite: | Solution NMR structures of proteins VPA0419 from Vibrio parahaemolyticus and yiiS from Shigella flexneri provide structural coverage for protein domain family PFAM 04175.
Proteins, 78, 2010
|
|
1ZCE
 
 | | X-Ray Crystal Structure of Protein Atu2648 from Agrobacterium tumefaciens. Northeast Structural Genomics Consortium Target AtR33. | | Descriptor: | hypothetical protein Atu2648 | | Authors: | Forouhar, F, Chen, Y, Conover, K, Acton, T.B, Montelione, G.T, Hunt, J.F, Tong, L, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2005-04-11 | | Release date: | 2005-04-19 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | Structural genomics reveals EVE as a new ASCH/PUA-related domain.
Proteins, 75, 2009
|
|
5EIY
 
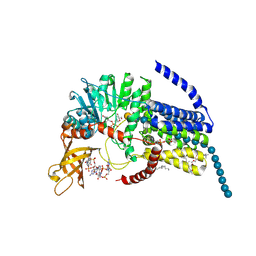 | | Bacterial cellulose synthase bound to a substrate analogue | | Descriptor: | 9,9'-[(2R,3R,3aS,5S,7aR,9R,10R,10aS,12S,14aR)-3,5,10,12-tetrahydroxy-5,12-dioxidooctahydro-2H,7H-difuro[3,2-d:3',2'-j][1,3,7,9,2,8]tetraoxadiphosphacyclododecine-2,9-diyl]bis(2-amino-1,9-dihydro-6H-purin-6-one), DIUNDECYL PHOSPHATIDYL CHOLINE, LAURYL DIMETHYLAMINE-N-OXIDE, ... | | Authors: | McNamara, J.T, Zimmer, J. | | Deposit date: | 2015-10-30 | | Release date: | 2016-03-09 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.95 Å) | | Cite: | Observing cellulose biosynthesis and membrane translocation in crystallo.
Nature, 531, 2016
|
|
5EJZ
 
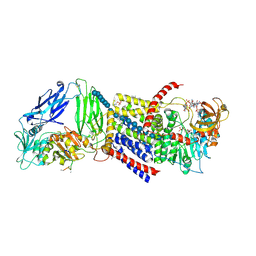 | | Bacterial Cellulose Synthase Product-Bound State | | Descriptor: | 1,2-Distearoyl-sn-glycerophosphoethanolamine, 2-deoxy-2-fluoro-beta-D-glucopyranose-(1-4)-beta-D-glucopyranose-(1-4)-beta-D-glucopyranose-(1-4)-beta-D-glucopyranose-(1-4)-beta-D-glucopyranose-(1-4)-beta-D-glucopyranose-(1-4)-beta-D-glucopyranose-(1-4)-beta-D-glucopyranose-(1-4)-beta-D-glucopyranose-(1-4)-beta-D-glucopyranose-(1-4)-beta-D-glucopyranose-(1-4)-beta-D-glucopyranose-(1-4)-beta-D-glucopyranose-(1-4)-beta-D-glucopyranose-(1-4)-beta-D-glucopyranose-(1-4)-beta-D-glucopyranose-(1-4)-beta-D-glucopyranose-(1-4)-beta-D-glucopyranose, 9,9'-[(2R,3R,3aS,5S,7aR,9R,10R,10aS,12S,14aR)-3,5,10,12-tetrahydroxy-5,12-dioxidooctahydro-2H,7H-difuro[3,2-d:3',2'-j][1,3,7,9,2,8]tetraoxadiphosphacyclododecine-2,9-diyl]bis(2-amino-1,9-dihydro-6H-purin-6-one), ... | | Authors: | Morgan, J.L.W, Zimmer, J. | | Deposit date: | 2015-11-02 | | Release date: | 2016-03-09 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.94 Å) | | Cite: | Observing cellulose biosynthesis and membrane translocation in crystallo.
Nature, 531, 2016
|
|
5EJ1
 
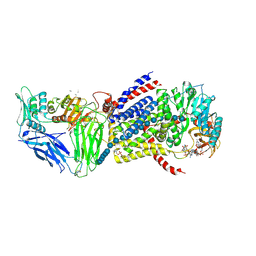 | | Pre-translocation state of bacterial cellulose synthase | | Descriptor: | (4S,7R)-7-(heptanoyloxy)-4-hydroxy-N,N,N-trimethyl-10-oxo-3,5,9-trioxa-4-phosphahexadecan-1-aminium 4-oxide, 1,2-DIACYL-SN-GLYCERO-3-PHOSPHOETHANOLAMINE, 9,9'-[(2R,3R,3aS,5S,7aR,9R,10R,10aS,12S,14aR)-3,5,10,12-tetrahydroxy-5,12-dioxidooctahydro-2H,7H-difuro[3,2-d:3',2'-j][1,3,7,9,2,8]tetraoxadiphosphacyclododecine-2,9-diyl]bis(2-amino-1,9-dihydro-6H-purin-6-one), ... | | Authors: | Moragn, J.L.W, Zimmer, J. | | Deposit date: | 2015-10-30 | | Release date: | 2016-03-30 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (3.4 Å) | | Cite: | Observing cellulose biosynthesis and membrane translocation in crystallo.
Nature, 531, 2016
|
|
2K3A
 
 | | NMR solution structure of Staphylococcus saprophyticus CHAP (cysteine, histidine-dependent amidohydrolases/peptidases) domain protein. Northeast Structural Genomics Consortium target SyR11 | | Descriptor: | CHAP domain protein | | Authors: | Rossi, P, Aramini, J.M, Chen, C.X, Nwosu, C, Cunningham, K.C, Owens, L.A, Xiao, R, Liu, J, Baran, M.C, Swapna, G, Acton, T.B, Rost, B, Montelione, G.T, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2008-04-29 | | Release date: | 2008-05-13 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Structural elucidation of the Cys-His-Glu-Asn proteolytic relay in the secreted CHAP domain enzyme from the human pathogen Staphylococcus saprophyticus.
Proteins, 74, 2008
|
|
5L0T
 
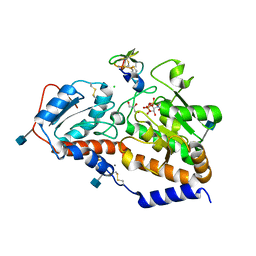 | | human POGLUT1 in complex with EGF(+) and UDP | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, CHLORIDE ION, ... | | Authors: | Li, Z, Rini, J.M. | | Deposit date: | 2016-07-28 | | Release date: | 2017-08-09 | | Last modified: | 2021-03-24 | | Method: | X-RAY DIFFRACTION (1.43 Å) | | Cite: | Structural basis of Notch O-glucosylation and O-xylosylation by mammalian protein-O-glucosyltransferase 1 (POGLUT1).
Nat Commun, 8, 2017
|
|
5L0U
 
 | | human POGLUT1 in complex with EGF(+) and UDP-phosphono-glucose | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, EGF(+), ... | | Authors: | Li, Z, Rini, J.M. | | Deposit date: | 2016-07-28 | | Release date: | 2017-08-09 | | Last modified: | 2021-03-24 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structural basis of Notch O-glucosylation and O-xylosylation by mammalian protein-O-glucosyltransferase 1 (POGLUT1).
Nat Commun, 8, 2017
|
|
5L0V
 
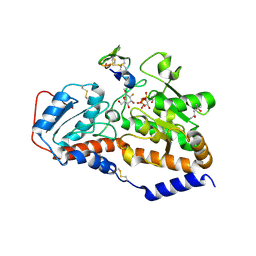 | | human POGLUT1 in complex with 2F-glucose modified EGF(+) and UDP | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-deoxy-2-fluoro-beta-D-glucopyranose, ... | | Authors: | Li, Z, Rini, J.M. | | Deposit date: | 2016-07-28 | | Release date: | 2017-08-09 | | Last modified: | 2021-03-24 | | Method: | X-RAY DIFFRACTION (1.305 Å) | | Cite: | Structural basis of Notch O-glucosylation and O-xylosylation by mammalian protein-O-glucosyltransferase 1 (POGLUT1).
Nat Commun, 8, 2017
|
|
5L0S
 
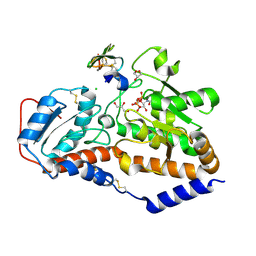 | | human POGLUT1 in complex with Factor VII EGF1 and UDP | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, CHLORIDE ION, ... | | Authors: | Li, Z, Rini, J.M. | | Deposit date: | 2016-07-28 | | Release date: | 2017-08-09 | | Last modified: | 2021-03-24 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | Structural basis of Notch O-glucosylation and O-xylosylation by mammalian protein-O-glucosyltransferase 1 (POGLUT1).
Nat Commun, 8, 2017
|
|
5L0R
 
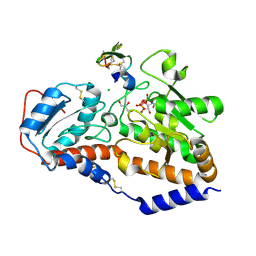 | | human POGLUT1 in complex with Notch1 EGF12 and UDP | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, CHLORIDE ION, ... | | Authors: | Li, Z, Rini, J.M. | | Deposit date: | 2016-07-28 | | Release date: | 2017-08-09 | | Last modified: | 2021-03-24 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Structural basis of Notch O-glucosylation and O-xylosylation by mammalian protein-O-glucosyltransferase 1 (POGLUT1).
Nat Commun, 8, 2017
|
|
6YRP
 
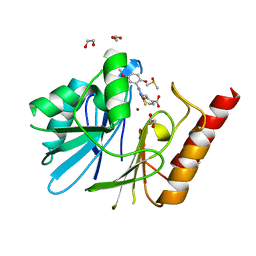 | | Crystal Structure of the VIM-2 Acquired Metallo-beta-Lactamase in Complex with JMV-4690 (Cpd 31) | | Descriptor: | 1,2-ETHANEDIOL, 2-[[[3-(5-methoxy-2-oxidanyl-phenyl)-5-sulfanylidene-1~{H}-1,2,4-triazol-4-yl]amino]methyl]benzoic acid, ACETATE ION, ... | | Authors: | Docquier, J.D, Pozzi, C, De Luca, F, Benvenuti, M, Mangani, S. | | Deposit date: | 2020-04-20 | | Release date: | 2020-09-30 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | 4-Amino-1,2,4-triazole-3-thione-derived Schiff bases as metallo-beta-lactamase inhibitors.
Eur.J.Med.Chem., 208, 2020
|
|
1IEZ
 
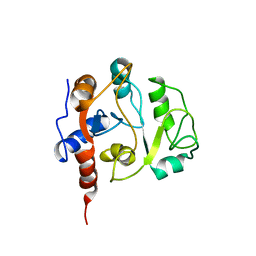 | | Solution Structure of 3,4-Dihydroxy-2-Butanone 4-Phosphate Synthase of Riboflavin Biosynthesis | | Descriptor: | 3,4-Dihydroxy-2-Butanone 4-Phosphate Synthase | | Authors: | Kelly, M.J.S, Ball, L.J, Kuhne, R, Bacher, A, Oschkinat, H. | | Deposit date: | 2001-04-11 | | Release date: | 2001-11-07 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | The NMR structure of the 47-kDa dimeric enzyme 3,4-dihydroxy-2-butanone-4-phosphate synthase and ligand binding studies reveal the location of the active site.
Proc.Natl.Acad.Sci.USA, 98, 2001
|
|
7LRK
 
 | | Crystal structure of BPTF bromodomain in complex with inhibitor Pdy-3-093 | | Descriptor: | 1,2-ETHANEDIOL, 4-chloranyl-2-methyl-5-[[(3~{S})-pyrrolidin-3-yl]amino]pyridazin-3-one, Nucleosome-remodeling factor subunit BPTF | | Authors: | Chan, A, Schonbrunn, E. | | Deposit date: | 2021-02-16 | | Release date: | 2022-08-10 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.44 Å) | | Cite: | New Design Rules for Developing Potent Cell-Active Inhibitors of the Nucleosome Remodeling Factor (NURF) via BPTF Bromodomain Inhibition.
J.Med.Chem., 64, 2021
|
|
7LP0
 
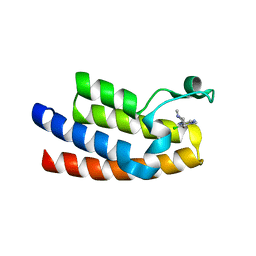 | | Crystal structure of BPTF bromodomain in complex with inhibitor Pdy-3-077 | | Descriptor: | 1,2-ETHANEDIOL, 4-chlorol-2-methyl-5-[[(3~{R})-1-methylpiperidin-3-yl]amino]pyridazin-3-one, DIMETHYL SULFOXIDE, ... | | Authors: | Chan, A, Schonbrunn, E. | | Deposit date: | 2021-02-11 | | Release date: | 2022-08-10 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.66 Å) | | Cite: | New Design Rules for Developing Potent Cell-Active Inhibitors of the Nucleosome Remodeling Factor (NURF) via BPTF Bromodomain Inhibition.
J.Med.Chem., 64, 2021
|
|
7LRO
 
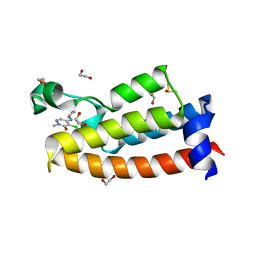 | | Crystal structure of BPTF bromodomain in complex with inhibitor HZ-01-105 | | Descriptor: | 1,2-ETHANEDIOL, 5-(azetidin-3-ylamino)-4-chloranyl-2-methyl-pyridazin-3-one, DIMETHYL SULFOXIDE, ... | | Authors: | Chan, A, Schonbrunn, E. | | Deposit date: | 2021-02-17 | | Release date: | 2022-08-10 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | New Design Rules for Developing Potent Cell-Active Inhibitors of the Nucleosome Remodeling Factor (NURF) via BPTF Bromodomain Inhibition.
J.Med.Chem., 64, 2021
|
|
7LPK
 
 | | Crystal structure of BPTF bromodomain in complex with inhibitor HZ-03-112 | | Descriptor: | 1,2-ETHANEDIOL, 4-chloranyl-2-methyl-5-[[(3~{R})-pyrrolidin-3-yl]amino]pyridazin-3-one, Nucleosome-remodeling factor subunit BPTF | | Authors: | Chan, A, Schonbrunn, E. | | Deposit date: | 2021-02-12 | | Release date: | 2022-08-10 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.39 Å) | | Cite: | New Design Rules for Developing Potent Cell-Active Inhibitors of the Nucleosome Remodeling Factor (NURF) via BPTF Bromodomain Inhibition.
J.Med.Chem., 64, 2021
|
|
4KP7
 
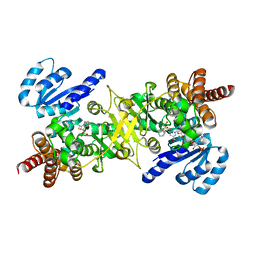 | | Structure of Plasmodium IspC in complex with a beta-thia-isostere derivative of Fosmidomycin | | Descriptor: | 1-deoxy-D-xylulose 5-phosphate reductoisomerase, apicoplast, MANGANESE (III) ION, ... | | Authors: | Kunfermann, A, Bacher, A, Groll, M. | | Deposit date: | 2013-05-13 | | Release date: | 2013-10-02 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | IspC as Target for Antiinfective Drug Discovery: Synthesis, Enantiomeric Separation, and Structural Biology of Fosmidomycin Thia Isosters.
J.Med.Chem., 56, 2013
|
|
2GGD
 
 | |
2GG4
 
 | | CP4 EPSP synthase (unliganded) | | Descriptor: | 3-phosphoshikimate 1-carboxyvinyltransferase | | Authors: | Schonbrunn, E, Funke, T. | | Deposit date: | 2006-03-23 | | Release date: | 2006-08-22 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Molecular basis for the herbicide resistance of Roundup Ready crops.
Proc.Natl.Acad.Sci.Usa, 103, 2006
|
|
2GGA
 
 | | CP4 EPSP synthase liganded with S3P and Glyphosate | | Descriptor: | 3-phosphoshikimate 1-carboxyvinyltransferase, GLYPHOSATE, SHIKIMATE-3-PHOSPHATE | | Authors: | Schonbrunn, E, Funke, T. | | Deposit date: | 2006-03-23 | | Release date: | 2006-08-22 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Molecular basis for the herbicide resistance of Roundup Ready crops.
Proc.Natl.Acad.Sci.Usa, 103, 2006
|
|
2GG6
 
 | | CP4 EPSP synthase liganded with S3P | | Descriptor: | 3-phosphoshikimate 1-carboxyvinyltransferase, SHIKIMATE-3-PHOSPHATE, SULFATE ION | | Authors: | Schonbrunn, E, Funke, T. | | Deposit date: | 2006-03-23 | | Release date: | 2006-08-22 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.64 Å) | | Cite: | Molecular basis for the herbicide resistance of Roundup Ready crops.
Proc.Natl.Acad.Sci.Usa, 103, 2006
|
|
1NAS
 
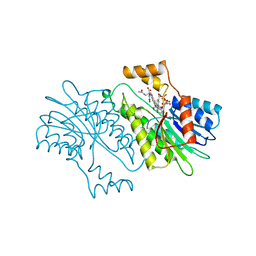 | | SEPIAPTERIN REDUCTASE COMPLEXED WITH N-ACETYL SEROTONIN | | Descriptor: | N-ACETYL SEROTONIN, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, OXALOACETATE ION, ... | | Authors: | Auerbach, G, Herrmann, A, Bacher, A, Huber, R. | | Deposit date: | 1998-03-26 | | Release date: | 1999-03-30 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | The 1.25 A crystal structure of sepiapterin reductase reveals its binding mode to pterins and brain neurotransmitters.
EMBO J., 16, 1997
|
|
