9FKB
 
 | | Tail of emppty Haloferax tailed virus 1 | | Descriptor: | Baseplate to tube adapter protein gp41, HK97 gp6-like/SPP1 gp15-like head-tail connector, MAGNESIUM ION, ... | | Authors: | Zhang, D, Daum, B, Isupov, M.N, McLaren, M. | | Deposit date: | 2024-06-03 | | Release date: | 2025-06-18 | | Method: | ELECTRON MICROSCOPY (2.96 Å) | | Cite: | CryoEM structure of Haloferax tailed virus
To Be Published
|
|
8OOD
 
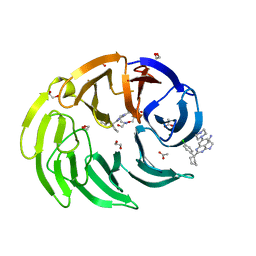 | |
8OO5
 
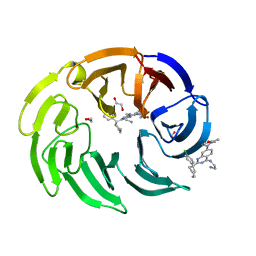 | |
8S2Y
 
 | | Crystal structure of alcohol oxidase PaAox1 from Picea abies | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, ... | | Authors: | Kruhler, N, Haataja, T, Sandgren, M. | | Deposit date: | 2024-02-19 | | Release date: | 2025-03-12 | | Method: | X-RAY DIFFRACTION (1.64 Å) | | Cite: | A glycerol oxidase from Norway spruce (Picea abies L. Karst.) expands biochemical and structural attributes of the GMC oxidoreductase superfamily
To Be Published
|
|
5J94
 
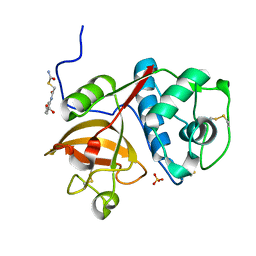 | | Human cathepsin K mutant C25S in complex with the allosteric effector NSC13345 | | Descriptor: | 2-{[(carbamoylsulfanyl)acetyl]amino}benzoic acid, Cathepsin K, SULFATE ION | | Authors: | Novinec, M, Korenc, M, Lenarcic, B, Baici, A. | | Deposit date: | 2016-04-08 | | Release date: | 2016-04-20 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.22002459 Å) | | Cite: | A novel allosteric mechanism in the cysteine peptidase cathepsin K discovered by computational methods.
Nat Commun, 5, 2014
|
|
6ZWT
 
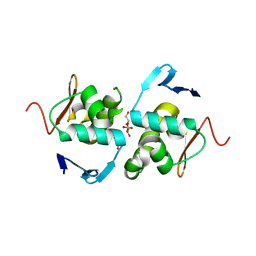 | | Crystal structure of DNA-binding domain of OmpR of two-component system of Acinetobacter baumannii | | Descriptor: | DI(HYDROXYETHYL)ETHER, GLYCEROL, SULFATE ION, ... | | Authors: | Narwal, M, Lucchini, V, Trebosc, V, Gartenmann, S, Pieren, M, Kemmer, C, Kammerer, R.A. | | Deposit date: | 2020-07-28 | | Release date: | 2021-08-11 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Targeting virulence regulation to disarm Acinetobacter baumannii pathogenesis.
Virulence, 13, 2022
|
|
8PX5
 
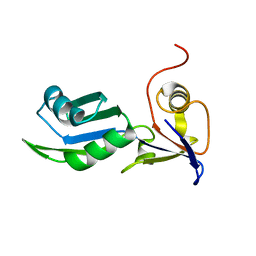 | | Structure of the RNA recognition motif (RRM) of Seb1 from S. pombe., solved at wavelength 2.75 A | | Descriptor: | Rpb7-binding protein seb1 | | Authors: | El Omari, K, Duman, R, Mykhaylyk, V, Orr, C, Wittmann, S, Renner, M, Grimes, J.M, Wagner, A. | | Deposit date: | 2023-07-22 | | Release date: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.77 Å) | | Cite: | Experimental phasing opportunities for macromolecular crystallography at very long wavelengths.
Commun Chem, 6, 2023
|
|
8QQJ
 
 | | CryoEM structure of the type IV pilin PilA5 from Thermus thermophilus | | Descriptor: | 7-Acetamido-5-acetimidoyl-3,5,7,9-tetradeoxy-L-glycero-L-manno-nonulosonic aci-(1-4)-alpha-D-mannopyranose-(1-3)-2-acetamido-2-deoxy-alpha-D-galactopyranose-(1-3)-2-acetamido-2-deoxy-alpha-D-galactopyranose, MAGNESIUM ION, Type IV narrow pilus major component PilA5 | | Authors: | Gold, V.A.M, Neuhaus, A, Gaines, M, Isupov, M, McLaren, M. | | Deposit date: | 2023-10-04 | | Release date: | 2024-10-16 | | Last modified: | 2024-11-20 | | Method: | ELECTRON MICROSCOPY (2.63 Å) | | Cite: | CryoEM structure of the type IV pilin PilA4 from Thermus thermophilus
To Be Published
|
|
5ULL
 
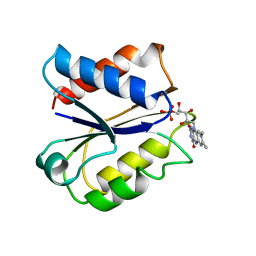 | | CLOSTRIDIUM BEIJERINCKII FLAVODOXIN: REDUCED | | Descriptor: | FLAVIN MONONUCLEOTIDE, FLAVODOXIN | | Authors: | Ludwig, M.L, Pattridge, K.A, Metzger, A.L, Dixon, M.M, Eren, M, Feng, Y, Swenson, R. | | Deposit date: | 1997-01-09 | | Release date: | 1997-03-12 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Control of oxidation-reduction potentials in flavodoxin from Clostridium beijerinckii: the role of conformation changes.
Biochemistry, 36, 1997
|
|
8QP0
 
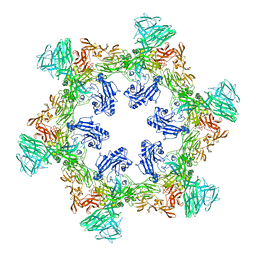 | | A hexamer pore in the S-layer of Sulfolobus acidocaldarius formed by SlaA protein | | Descriptor: | S-layer protein A | | Authors: | Gambelli, L, McLaren, M, Isupov, M, Conners, R, Daum, B. | | Deposit date: | 2023-09-29 | | Release date: | 2024-02-21 | | Last modified: | 2024-11-13 | | Method: | ELECTRON MICROSCOPY (11.2 Å) | | Cite: | Structure of the two-component S-layer of the archaeon Sulfolobus acidocaldarius.
Elife, 13, 2024
|
|
8QOX
 
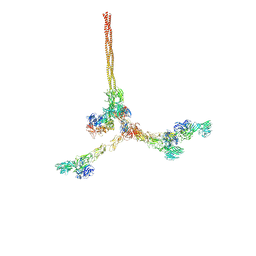 | | Two-component assembly of SlaA and SlaB S-layer proteins of Sulfolobus acidocaldarius | | Descriptor: | Conserved membrane protein, S-layer protein A | | Authors: | Gambelli, L, McLaren, M, Isupov, M, Conners, R, Daum, B. | | Deposit date: | 2023-09-29 | | Release date: | 2024-02-21 | | Last modified: | 2024-11-13 | | Method: | ELECTRON MICROSCOPY (11.2 Å) | | Cite: | Structure of the two-component S-layer of the archaeon Sulfolobus acidocaldarius.
Elife, 13, 2024
|
|
8PDP
 
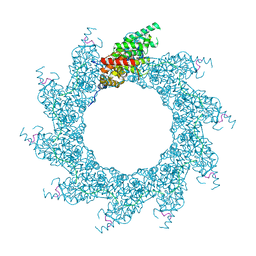 | | 10-mer ring of HMPV N-RNA bound to the C-terminal region of P | | Descriptor: | Nucleoprotein, Phosphoprotein, RNA | | Authors: | Whitehead, J.D, Decool, H, Leyrat, C, Carrique, L, Fix, J, Eleouet, J.F, Galloux, M, Renner, M. | | Deposit date: | 2023-06-12 | | Release date: | 2023-12-06 | | Last modified: | 2024-03-20 | | Method: | ELECTRON MICROSCOPY (2.9 Å) | | Cite: | Structure of the N-RNA/P interface indicates mode of L/P recruitment to the nucleocapsid of human metapneumovirus.
Nat Commun, 14, 2023
|
|
8PDM
 
 | | 11-mer ring of human metapneumovirus (HMPV) N-RNA | | Descriptor: | Nucleoprotein, RNA | | Authors: | Whitehead, J.D, Decool, H, Leyrat, C, Carrique, L, Fix, J, Eleouet, J.F, Galloux, M, Renner, M. | | Deposit date: | 2023-06-12 | | Release date: | 2023-12-06 | | Last modified: | 2024-03-20 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Structure of the N-RNA/P interface indicates mode of L/P recruitment to the nucleocapsid of human metapneumovirus.
Nat Commun, 14, 2023
|
|
8PDN
 
 | | Spiral of assembled human metapneumovirus (HMPV) N-RNA | | Descriptor: | Nucleoprotein, RNA | | Authors: | Whitehead, J.D, Decool, H, Leyrat, C, Carrique, L, Fix, J, Eleouet, J.F, Galloux, M, Renner, M. | | Deposit date: | 2023-06-12 | | Release date: | 2023-12-06 | | Last modified: | 2025-07-09 | | Method: | ELECTRON MICROSCOPY (4.7 Å) | | Cite: | Structure of the N-RNA/P interface indicates mode of L/P recruitment to the nucleocapsid of human metapneumovirus.
Nat Commun, 14, 2023
|
|
8PDQ
 
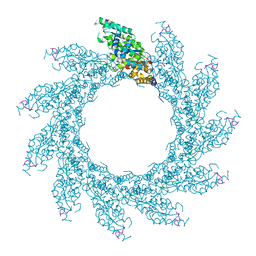 | | 11-mer ring of HMPV N-RNA bound to the C-terminal region of P | | Descriptor: | Nucleoprotein, Phosphoprotein, RNA | | Authors: | Whitehead, J.D, Decool, H, Leyrat, C, Carrique, L, Fix, J, Eleouet, J.F, Galloux, M, Renner, M. | | Deposit date: | 2023-06-12 | | Release date: | 2023-12-06 | | Last modified: | 2024-03-20 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Structure of the N-RNA/P interface indicates mode of L/P recruitment to the nucleocapsid of human metapneumovirus.
Nat Commun, 14, 2023
|
|
8PDR
 
 | | Rigid body fit of assembled HMPV N-RNA spiral bound to the C-terminal region of P | | Descriptor: | Nucleoprotein, Phosphoprotein, RNA | | Authors: | Whitehead, J.D, Decool, H, Leyrat, C, Carrique, L, Fix, J, Eleouet, J.F, Galloux, M, Renner, M. | | Deposit date: | 2023-06-12 | | Release date: | 2023-12-06 | | Last modified: | 2025-07-02 | | Method: | ELECTRON MICROSCOPY (4 Å) | | Cite: | Structure of the N-RNA/P interface indicates mode of L/P recruitment to the nucleocapsid of human metapneumovirus.
Nat Commun, 14, 2023
|
|
8PDO
 
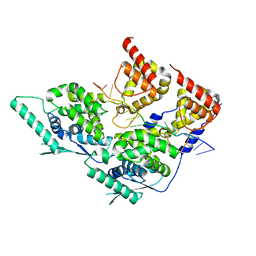 | | Local refinement of dimeric human metapneumovirus (HMPV) N-RNA | | Descriptor: | Nucleoprotein, RNA | | Authors: | Whitehead, J.D, Decool, H, Leyrat, C, Carrique, L, Fix, J, Eleouet, J.F, Galloux, M, Renner, M. | | Deposit date: | 2023-06-12 | | Release date: | 2023-12-06 | | Last modified: | 2024-03-20 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Structure of the N-RNA/P interface indicates mode of L/P recruitment to the nucleocapsid of human metapneumovirus.
Nat Commun, 14, 2023
|
|
8PDL
 
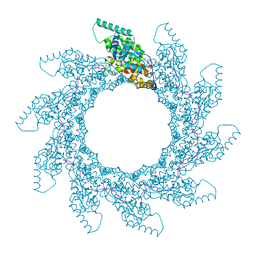 | | 10-mer ring of human metapneumovirus (HMPV) N-RNA | | Descriptor: | Nucleoprotein, RNA | | Authors: | Whitehead, J.D, Decool, H, Leyrat, C, Carrique, L, Fix, J, Eleouet, J.F, Galloux, M, Renner, M. | | Deposit date: | 2023-06-12 | | Release date: | 2023-12-06 | | Last modified: | 2024-03-20 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Structure of the N-RNA/P interface indicates mode of L/P recruitment to the nucleocapsid of human metapneumovirus.
Nat Commun, 14, 2023
|
|
8PDS
 
 | | Local refinement of dimeric HMPV N-RNA bound to the C-terminal region of P | | Descriptor: | Nucleoprotein, Phosphoprotein, RNA | | Authors: | Whitehead, J.D, Decool, H, Leyrat, C, Carrique, L, Fix, J, Eleouet, J.F, Galloux, M, Renner, M. | | Deposit date: | 2023-06-12 | | Release date: | 2023-12-06 | | Last modified: | 2024-03-20 | | Method: | ELECTRON MICROSCOPY (2.9 Å) | | Cite: | Structure of the N-RNA/P interface indicates mode of L/P recruitment to the nucleocapsid of human metapneumovirus.
Nat Commun, 14, 2023
|
|
5FOH
 
 | | Crystal structure of the catalytic domain of NcLPMO9A | | Descriptor: | COPPER (II) ION, LITHIUM ION, POLYSACCHARIDE MONOOXYGENASE, ... | | Authors: | Westereng, B, Kracun, S.K, Dimarogona, M, Mathiesen, G, Willats, W.G.T, Sandgren, M, Aachmann, F.L, Eijsink, V.G.H. | | Deposit date: | 2015-11-18 | | Release date: | 2016-12-07 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Comparison of three seemingly similar lytic polysaccharide monooxygenases fromNeurospora crassasuggests different roles in plant biomass degradation.
J.Biol.Chem., 2019
|
|
8Q30
 
 | | Sulfolobus acidocaldarius AAP filament. | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 6-deoxy-6-sulfo-beta-D-glucopyranose-(1-3)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Isupov, M.N, Gaines, M, Daum, B, McLaren, M. | | Deposit date: | 2023-08-03 | | Release date: | 2024-03-06 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (3.22 Å) | | Cite: | CryoEM reveals the structure of an archaeal pilus involved in twitching motility.
Nat Commun, 15, 2024
|
|
5OSI
 
 | | Structure of retromer VPS29-VPS35C subunits complexed with RidL harpin loop (163-176) | | Descriptor: | 1,2-ETHANEDIOL, Interaptin, SODIUM ION, ... | | Authors: | Romano-Moreno, M, Rojas, A.L, Lucas, M, Isupov, M.N, Hierro, A. | | Deposit date: | 2017-08-17 | | Release date: | 2017-12-13 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.52 Å) | | Cite: | Molecular mechanism for the subversion of the retromer coat by the Legionella effector RidL.
Proc. Natl. Acad. Sci. U.S.A., 114, 2017
|
|
5OSH
 
 | | Structure of retromer VPS29-VPS35C subunits complexed with RidL N-terminal domain (1-236) | | Descriptor: | Interaptin, Vacuolar protein sorting-associated protein 29, Vacuolar protein sorting-associated protein 35 | | Authors: | Romano-Moreno, M, Rojas, A.L, Lucas, M, Isupov, M.N, Hierro, A. | | Deposit date: | 2017-08-17 | | Release date: | 2017-12-13 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (4.3 Å) | | Cite: | Molecular mechanism for the subversion of the retromer coat by the Legionella effector RidL.
Proc. Natl. Acad. Sci. U.S.A., 114, 2017
|
|
8QRD
 
 | | OleP in complex with testosterone in high salt crystallization conditions | | Descriptor: | Cytochrome P-450, FORMIC ACID, PROTOPORPHYRIN IX CONTAINING FE, ... | | Authors: | Fata, F, Costanzo, A, Freda, I, Gugole, E, Bulfaro, G, Barbizzi, L, Di Renzo, M, Savino, C, Vallone, B, Montemiglio, L.C. | | Deposit date: | 2023-10-06 | | Release date: | 2024-07-03 | | Last modified: | 2025-01-22 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Binding of steroid substrates reveals the key to the productive transition of the cytochrome P450 OleP.
Structure, 32, 2024
|
|
8QYI
 
 | | OleP in complex with lithocholic acid in high salt crystallization conditions | | Descriptor: | (3beta,5beta,14beta,17alpha)-3-hydroxycholan-24-oic acid, Cytochrome P-450, FORMIC ACID, ... | | Authors: | Fata, F, Costanzo, A, Freda, I, Gugole, E, Bulfaro, G, Barbizzi, L, Di Renzo, M, Savino, C, Vallone, B, Montemiglio, L.C. | | Deposit date: | 2023-10-26 | | Release date: | 2024-07-03 | | Last modified: | 2025-01-22 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Binding of steroid substrates reveals the key to the productive transition of the cytochrome P450 OleP.
Structure, 32, 2024
|
|
