8OHG
 
 | | PanDDA analysis group deposition -- CdaA in complex with fragment F2X-Entry F09 | | Descriptor: | 4-pyridin-2-ylphenol, Cyclic di-AMP synthase CdaA, MAGNESIUM ION | | Authors: | Garbers, T.B, Neumann, P, Wollenhaupt, J, Weiss, M.S, Ficner, R. | | Deposit date: | 2023-03-21 | | Release date: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.25 Å) | | Cite: | PanDDA analysis group deposition -- CdaA in complex with fragment F2X-Entry F09
To Be Published
|
|
8OHE
 
 | | PanDDA analysis group deposition -- CdaA in complex with fragment F2X-Entry F03 | | Descriptor: | Cyclic di-AMP synthase CdaA, MAGNESIUM ION, N-(3-fluorophenyl)-2-(2-methoxyethoxy)acetamide | | Authors: | Garbers, T.B, Neumann, P, Wollenhaupt, J, Weiss, M.S, Ficner, R. | | Deposit date: | 2023-03-21 | | Release date: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.15 Å) | | Cite: | PanDDA analysis group deposition -- CdaA in complex with fragment F2X-Entry F03
To Be Published
|
|
8OHC
 
 | | PanDDA analysis group deposition -- CdaA in complex with fragment F2X-Entry E12 | | Descriptor: | 6-azanyl-3-methyl-1,3-benzoxazol-2-one, Cyclic di-AMP synthase CdaA, MAGNESIUM ION | | Authors: | Garbers, T.B, Neumann, P, Wollenhaupt, J, Weiss, M.S, Ficner, R. | | Deposit date: | 2023-03-21 | | Release date: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.17 Å) | | Cite: | PanDDA analysis group deposition -- CdaA in complex with fragment F2X-Entry E12
To Be Published
|
|
3NS6
 
 | |
3NS5
 
 | |
3OGZ
 
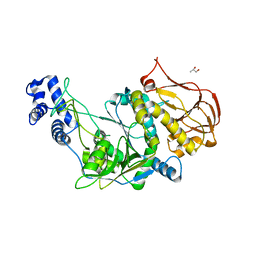 | | Protein structure of USP from L. major in Apo-form | | Descriptor: | GLYCEROL, UDP-sugar pyrophosphorylase | | Authors: | Dickmanns, A, Damerow, S, Neumann, P, Schulz, E.-C, Lamerz, A, Routier, F, Ficner, R. | | Deposit date: | 2010-08-17 | | Release date: | 2010-11-17 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.03 Å) | | Cite: | Structural basis for the broad substrate range of the UDP-sugar pyrophosphorylase from Leishmania major.
J.Mol.Biol., 405, 2011
|
|
3OE1
 
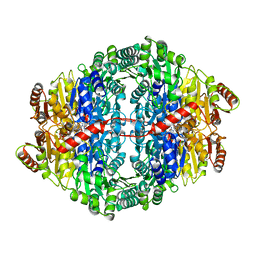 | | Pyruvate decarboxylase variant Glu473Asp from Z. mobilis in complex with reaction intermediate 2-lactyl-ThDP | | Descriptor: | 3-[(4-AMINO-2-METHYLPYRIMIDIN-5-YL)METHYL]-2-(1-CARBOXY-1-HYDROXYETHYL)-5-(2-{[HYDROXY(PHOSPHONOOXY)PHOSPHORYL]OXY}ETHYL)-4-METHYL-1,3-THIAZOL-3-IUM, GLYCEROL, MAGNESIUM ION, ... | | Authors: | Meyer, D, Neumann, P, Parthier, C, Tittmann, K. | | Deposit date: | 2010-08-12 | | Release date: | 2010-09-08 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.985 Å) | | Cite: | Double duty for a conserved glutamate in pyruvate decarboxylase: evidence of the participation in stereoelectronically controlled decarboxylation and in protonation of the nascent carbanion/enamine intermediate .
Biochemistry, 49, 2010
|
|
3OH2
 
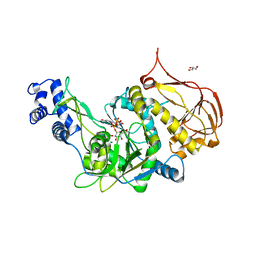 | | Protein structure of USP from L. major bound to URIDINE-5'-DIPHOSPHATE-GALACTOSE | | Descriptor: | GALACTOSE-URIDINE-5'-DIPHOSPHATE, GLYCEROL, UDP-sugar pyrophosphorylase | | Authors: | Dickmanns, A, Damerow, S, Neumann, P, Schulz, E.-C, Lamerz, A, Routier, F, Ficner, R. | | Deposit date: | 2010-08-17 | | Release date: | 2010-11-17 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.14 Å) | | Cite: | Structural basis for the broad substrate range of the UDP-sugar pyrophosphorylase from Leishmania major.
J.Mol.Biol., 405, 2011
|
|
3OH3
 
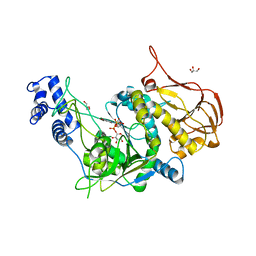 | | Protein structure of USP from L. major bound to URIDINE-5'-DIPHOSPHATE -Arabinose | | Descriptor: | GLYCEROL, UDP-sugar pyrophosphorylase, [(2R,3S,4R,5R)-5-(2,4-dioxo-3,4-dihydropyrimidin-1(2H)-yl)-3,4-dihydroxytetrahydrofuran-2-yl]methyl (2S,3R,4S,5S)-3,4,5-trihydroxytetrahydro-2H-pyran-2-yl dihydrogen diphosphate | | Authors: | Dickmanns, A, Damerow, S, Neumann, P, Schulz, E.-C, Lamerz, A, Routier, F, Ficner, R. | | Deposit date: | 2010-08-17 | | Release date: | 2010-11-17 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.03 Å) | | Cite: | Structural basis for the broad substrate range of the UDP-sugar pyrophosphorylase from Leishmania major.
J.Mol.Biol., 405, 2011
|
|
3OH4
 
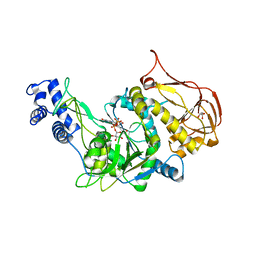 | | Protein structure of USP from L. major bound to URIDINE-5'-DIPHOSPHATE Glucose | | Descriptor: | GLYCEROL, UDP-sugar pyrophosphorylase, URIDINE-5'-DIPHOSPHATE-GLUCOSE | | Authors: | Dickmanns, A, Damerow, S, Neumann, P, Schulz, E.-C, Lamerz, A, Routier, F.H, Ficner, R. | | Deposit date: | 2010-08-17 | | Release date: | 2010-11-17 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.21 Å) | | Cite: | Structural basis for the broad substrate range of the UDP-sugar pyrophosphorylase from Leishmania major.
J.Mol.Biol., 405, 2011
|
|
3OH1
 
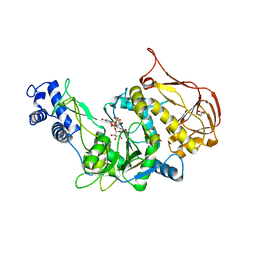 | | Protein structure of USP from L. major bound to URIDINE-5'-DIPHOSPHATE-Galacturonic acid | | Descriptor: | (2S,3R,4S,5R,6R)-6-[[[(2R,3S,4R,5R)-5-(2,4-dioxopyrimidin-1-yl)-3,4-dihydroxy-oxolan-2-yl]methoxy-hydroxy-phosphoryl]oxy-hydroxy-phosphoryl]oxy-3,4,5-trihydroxy-oxane-2-carboxylic acid, GLYCEROL, UDP-sugar pyrophosphorylase | | Authors: | Dickmanns, A, Damerow, S, Neumann, P, Schulz, E.-C, Lamerz, A, Routier, F, Ficner, R. | | Deposit date: | 2010-08-17 | | Release date: | 2010-11-17 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.18 Å) | | Cite: | Structural basis for the broad substrate range of the UDP-sugar pyrophosphorylase from Leishmania major.
J.Mol.Biol., 405, 2011
|
|
3OH0
 
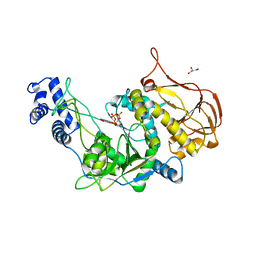 | | Protein structure of USP from L. major bound to URIDINE-5'-TRIPHOSPHATE | | Descriptor: | GLYCEROL, UDP-sugar pyrophosphorylase, URIDINE 5'-TRIPHOSPHATE | | Authors: | Dickmanns, A, Damerow, S, Neumann, P, Schulz, E.-C, Lamerz, A, Routier, F, Ficner, R. | | Deposit date: | 2010-08-17 | | Release date: | 2010-11-17 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Structural basis for the broad substrate range of the UDP-sugar pyrophosphorylase from Leishmania major.
J.Mol.Biol., 405, 2011
|
|
3S1U
 
 | | Transaldolase from Thermoplasma acidophilum in complex with D-erythrose 4-phosphate | | Descriptor: | CHLORIDE ION, ERYTHOSE-4-PHOSPHATE, Probable transaldolase | | Authors: | Lehwess-Litzmann, A, Neumann, P, Parthier, C, Tittmann, K. | | Deposit date: | 2011-05-16 | | Release date: | 2011-08-24 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Twisted Schiff base intermediates and substrate locale revise transaldolase mechanism.
Nat.Chem.Biol., 7, 2011
|
|
3S1X
 
 | | Transaldolase from Thermoplasma acidophilum in complex with D-sedoheptulose 7-phosphate Schiff-base intermediate | | Descriptor: | D-ALTRO-HEPT-2-ULOSE 7-PHOSPHATE, Probable transaldolase | | Authors: | Lehwess-Litzmann, A, Neumann, P, Parthier, C, Tittmann, K. | | Deposit date: | 2011-05-16 | | Release date: | 2011-08-24 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Twisted Schiff base intermediates and substrate locale revise transaldolase mechanism.
Nat.Chem.Biol., 7, 2011
|
|
3S0C
 
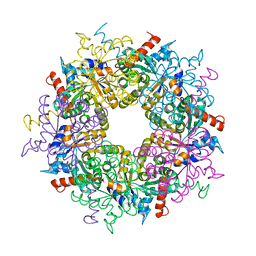 | | Transaldolase wt of Thermoplasma acidophilum | | Descriptor: | GLYCEROL, Probable transaldolase | | Authors: | Lehwess-Litzmann, A, Neumann, P, Parthier, C, Tittmann, K. | | Deposit date: | 2011-05-13 | | Release date: | 2011-08-24 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.78 Å) | | Cite: | Twisted Schiff base intermediates and substrate locale revise transaldolase mechanism.
Nat.Chem.Biol., 7, 2011
|
|
3S1W
 
 | | Transaldolase variant Lys86Ala from Thermoplasma acidophilum in complex with glycerol and citrate | | Descriptor: | CITRATE ANION, GLYCEROL, Probable transaldolase | | Authors: | Lehwess-Litzmann, A, Neumann, P, Parthier, C, Tittmann, K. | | Deposit date: | 2011-05-16 | | Release date: | 2011-08-24 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Twisted Schiff base intermediates and substrate locale revise transaldolase mechanism.
Nat.Chem.Biol., 7, 2011
|
|
4XRI
 
 | | Crystal Structure of Importin Beta in an Ammonium Sulfate Condition | | Descriptor: | GLYCEROL, Putative uncharacterized protein, SULFATE ION | | Authors: | Tauchert, M.J, Neumann, P, Ficner, R, Dickmanns, A. | | Deposit date: | 2015-01-21 | | Release date: | 2016-01-27 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Impact of the crystallization condition on importin-beta conformation.
Acta Crystallogr D Struct Biol, 72, 2016
|
|
4XRK
 
 | |
4ZP1
 
 | | Crystal structure of Zymomonas mobilis pyruvate decarboxylase variant Glu473Ala | | Descriptor: | GLYCEROL, MAGNESIUM ION, NICKEL (II) ION, ... | | Authors: | Wechsler, C, Neumann, P, Tittmann, K. | | Deposit date: | 2015-05-07 | | Release date: | 2015-11-04 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.205 Å) | | Cite: | Tuning and Switching Enantioselectivity of Asymmetric Carboligation in an Enzyme through Mutational Analysis of a Single Hot Spot.
Chembiochem, 16, 2015
|
|
2FYI
 
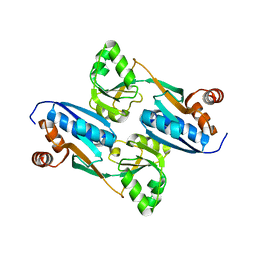 | | Crystal Structure of the Cofactor-Binding Domain of the Cbl Transcriptional Regulator | | Descriptor: | HTH-type transcriptional regulator cbl | | Authors: | Stec, E, Neumann, P, Wilkinson, A.J, Brzozowski, A.M, Bujacz, G.D. | | Deposit date: | 2006-02-08 | | Release date: | 2006-02-21 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structural Basis of the Sulphate Starvation Response in E. coli: Crystal Structure and Mutational Analysis of the Cofactor-binding Domain of the Cbl Transcriptional Regulator.
J.Mol.Biol., 364, 2006
|
|
5AFI
 
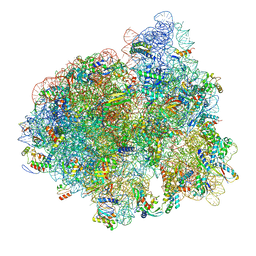 | | 2.9A Structure of E. coli ribosome-EF-TU complex by cs-corrected cryo-EM | | Descriptor: | 16S ribosomal RNA, 23S ribosomal RNA, 30S ribosomal protein S10, ... | | Authors: | Fischer, N, Neumann, P, Konevega, A.L, Bock, L.V, Ficner, R, Rodnina, M.V, Stark, H. | | Deposit date: | 2015-01-22 | | Release date: | 2015-03-11 | | Last modified: | 2024-04-24 | | Method: | ELECTRON MICROSCOPY (2.9 Å) | | Cite: | Structure of the E. coli ribosome-EF-Tu complex at <3 angstrom resolution by Cs-corrected cryo-EM.
Nature, 520, 2015
|
|
6TVO
 
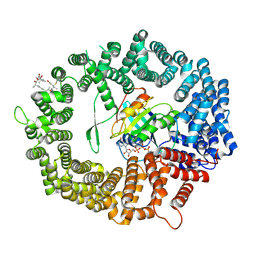 | | Human CRM1-RanGTP in complex with Leptomycin B | | Descriptor: | Exportin-1, GTP-binding nuclear protein Ran, GUANOSINE-5'-TRIPHOSPHATE, ... | | Authors: | Shaikhqasem, A, Ficner, R. | | Deposit date: | 2020-01-10 | | Release date: | 2020-07-08 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (3.201 Å) | | Cite: | Characterization of Inhibition Reveals Distinctive Properties for Human andSaccharomyces cerevisiaeCRM1.
J.Med.Chem., 63, 2020
|
|
3NOL
 
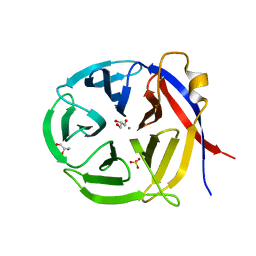 | | Crystal structure of Zymomonas mobilis Glutaminyl Cyclase (trigonal form) | | Descriptor: | CALCIUM ION, GLYCEROL, Glutamine cyclotransferase, ... | | Authors: | Parthier, C, Carrillo, D.R, Stubbs, M.T. | | Deposit date: | 2010-06-25 | | Release date: | 2010-11-03 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Kinetic and structural characterization of bacterial glutaminyl cyclases from Zymomonas mobilis and Myxococcus xanthus
Biol.Chem., 391, 2010
|
|
6ZM2
 
 | |
6SUR
 
 | | The Rab33B-Atg16L1 crystal structure | | Descriptor: | Autophagy-related protein 16-1, GUANOSINE-5'-TRIPHOSPHATE, MAGNESIUM ION, ... | | Authors: | Metje-Sprink, J, Kuehnel, K. | | Deposit date: | 2019-09-16 | | Release date: | 2020-08-05 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (3.467 Å) | | Cite: | Crystal structure of the Rab33B/Atg16L1 effector complex.
Sci Rep, 10, 2020
|
|
