7X2A
 
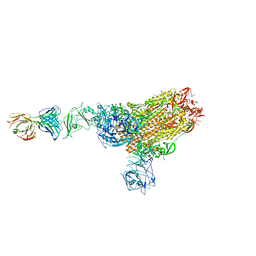 | | MERS-CoV spike complex with S41 neutralizing antibody Fab Class1 (1u2d RBD with 1Fab) | | Descriptor: | MERS-CoV Spike glycoprotein, antibody S41 heavy chain, antibody S41 light chain | | Authors: | Zeng, J.W, Zhang, S.Y, Zhou, H.X, Wang, X.W. | | Deposit date: | 2022-02-25 | | Release date: | 2022-11-09 | | Last modified: | 2022-11-23 | | Method: | ELECTRON MICROSCOPY (2.49 Å) | | Cite: | Cryoelectron microscopy structures of a human neutralizing antibody bound to MERS-CoV spike glycoprotein.
Front Microbiol, 13, 2022
|
|
7X26
 
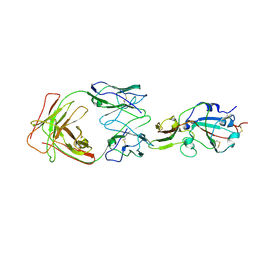 | | S41 neutralizing antibody Fab(MERS-CoV) | | Descriptor: | Spike glycoprotein, antibody S41 heavy chain, antibody S41 light chain | | Authors: | Zeng, J.W, Zhang, S.Y, Wang, X.W. | | Deposit date: | 2022-02-25 | | Release date: | 2022-11-09 | | Last modified: | 2022-11-23 | | Method: | ELECTRON MICROSCOPY (3.685 Å) | | Cite: | Cryoelectron microscopy structures of a human neutralizing antibody bound to MERS-CoV spike glycoprotein.
Front Microbiol, 13, 2022
|
|
7X29
 
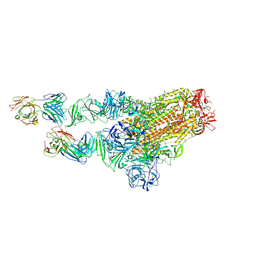 | | MERS-CoV spike complex with S41 neutralizing antibody Fab Class2 (1u2d RBD with 2Fab) | | Descriptor: | Spike glycoprotein, antibody S41 heavy chain, antibody S41 light chain | | Authors: | Zeng, J.W, Zhang, S.Y, Zhou, H.X, Wang, X.W. | | Deposit date: | 2022-02-25 | | Release date: | 2022-11-09 | | Last modified: | 2022-11-23 | | Method: | ELECTRON MICROSCOPY (2.49 Å) | | Cite: | Cryoelectron microscopy structures of a human neutralizing antibody bound to MERS-CoV spike glycoprotein.
Front Microbiol, 13, 2022
|
|
2Q8Z
 
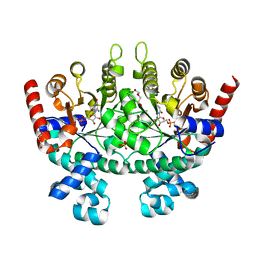 | | Crystal structure of Plasmodium falciparum orotidine 5'-phosphate decarboxylase complexed with 6-amino-UMP | | Descriptor: | 2-(2-(2-(2-(2-(2-ETHOXYETHOXY)ETHOXY)ETHOXY)ETHOXY)ETHOXY)ETHANOL, 6-AMINOURIDINE 5'-MONOPHOSPHATE, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Liu, Y, Lau, W, Bello, A.M, Kotra, L.P, Hui, R, Pai, E.F. | | Deposit date: | 2007-06-12 | | Release date: | 2008-01-29 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structure-Activity Relationships of C6-Uridine Derivatives Targeting Plasmodia Orotidine Monophosphate Decarboxylase.
J.Med.Chem., 51, 2008
|
|
7X28
 
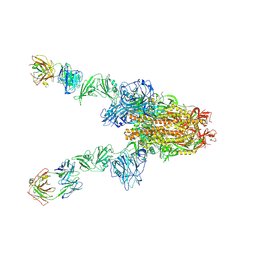 | | MERS-CoV spike complex with S41 neutralizing antibody Fab Class3 (2u1d RBD with 2Fab) | | Descriptor: | Spike glycoprotein, antibody S41 heavy chain, antibody S41 light chain | | Authors: | Zeng, J.W, Zhang, S.Y, Zhou, H.X, Wang, X.W. | | Deposit date: | 2022-02-25 | | Release date: | 2023-01-18 | | Last modified: | 2023-01-25 | | Method: | ELECTRON MICROSCOPY (2.49 Å) | | Cite: | Cryoelectron microscopy structures of a human neutralizing antibody bound to MERS-CoV spike glycoprotein.
Front Microbiol, 13, 2022
|
|
2XSP
 
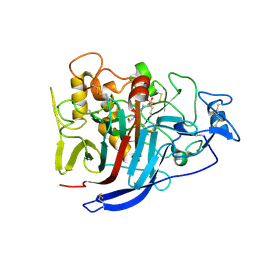 | | Structure of Cellobiohydrolase 1 (Cel7A) from Heterobasidion annosum | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, CELLULOSE 1,4-BETA-CELLOBIOSIDASE, ... | | Authors: | Haddad-momeni, M, Hansson, H, Mikkelsen, N.E, Wang, X, Svedberg, J, Sandgren, M, Stahlberg, J. | | Deposit date: | 2010-09-29 | | Release date: | 2011-10-12 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structural, Biochemical, and Computational Characterization of the Glycoside Hydrolase Family 7 Cellobiohydrolase of the Tree-Killing Fungus Heterobasidion Irregulare.
J.Biol.Chem., 288, 2013
|
|
2LNV
 
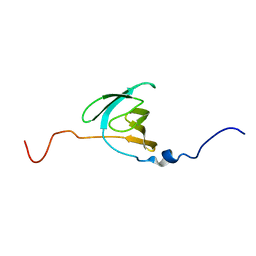 | |
2QF6
 
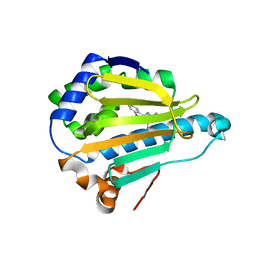 | | HSP90 complexed with A56322 | | Descriptor: | 6-(3-BROMO-2-NAPHTHYL)-1,3,5-TRIAZINE-2,4-DIAMINE, Heat shock protein HSP 90-alpha | | Authors: | Park, C.H. | | Deposit date: | 2007-06-26 | | Release date: | 2008-07-01 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Discovery and design of novel HSP90 inhibitors using multiple fragment-based design strategies.
Chem.Biol.Drug Des., 70, 2007
|
|
2QG2
 
 | | HSP90 complexed with A917985 | | Descriptor: | 3-({2-[(2-AMINO-6-METHYLPYRIMIDIN-4-YL)ETHYNYL]BENZYL}AMINO)-1,3-OXAZOL-2(3H)-ONE, Heat shock protein HSP 90-alpha | | Authors: | Park, C.H. | | Deposit date: | 2007-06-28 | | Release date: | 2008-07-01 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Discovery and design of novel HSP90 inhibitors using multiple fragment-based design strategies.
Chem.Biol.Drug Des., 70, 2007
|
|
2MVH
 
 | |
2QAF
 
 | | Crystal structure of Plasmodium falciparum orotidine 5'-phosphate decarboxylase covalently modified by 6-iodo-UMP | | Descriptor: | Orotidine 5' monophosphate decarboxylase, SULFATE ION, URIDINE-5'-MONOPHOSPHATE | | Authors: | Liu, Y, Lau, W, Bello, A.M, Kotra, L.P, Hui, R, Pai, E.F. | | Deposit date: | 2007-06-15 | | Release date: | 2008-04-29 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Structure-activity relationships of C6-uridine derivatives targeting plasmodia orotidine monophosphate decarboxylase
J.Med.Chem., 51, 2008
|
|
2QG0
 
 | | HSP90 complexed with A943037 | | Descriptor: | Heat shock protein HSP 90-alpha, N-[(2-AMINO-6-METHYLPYRIMIDIN-4-YL)METHYL]-3-{[(E)-(2-OXODIHYDROFURAN-3(2H)-YLIDENE)METHYL]AMINO}BENZENESULFONAMIDE | | Authors: | Park, C.H. | | Deposit date: | 2007-06-28 | | Release date: | 2008-07-01 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Discovery and design of novel HSP90 inhibitors using multiple fragment-based design strategies.
Chem.Biol.Drug Des., 70, 2007
|
|
2QFO
 
 | | HSP90 complexed with A143571 and A516383 | | Descriptor: | (3E)-3-[(phenylamino)methylidene]dihydrofuran-2(3H)-one, 4-METHYL-6-(TRIFLUOROMETHYL)PYRIMIDIN-2-AMINE, Heat shock protein HSP 90-alpha | | Authors: | Park, C.H. | | Deposit date: | 2007-06-27 | | Release date: | 2008-07-01 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.68 Å) | | Cite: | Discovery and design of novel HSP90 inhibitors using multiple fragment-based design strategies.
CHEM.BIOL.DRUG DES., 70, 2007
|
|
5XNB
 
 | |
5X38
 
 | |
5X35
 
 | |
5X36
 
 | |
5X37
 
 | |
5X3C
 
 | |
7WL3
 
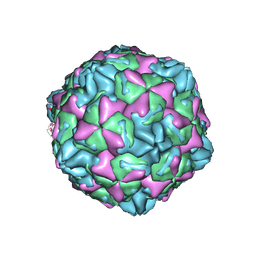 | | CVB5 expended empty particle | | Descriptor: | Capsid protein, Genome polyprotein | | Authors: | Yang, P, Wang, K. | | Deposit date: | 2022-01-12 | | Release date: | 2022-03-30 | | Last modified: | 2024-06-26 | | Method: | ELECTRON MICROSCOPY (2.95 Å) | | Cite: | Atomic Structures of Coxsackievirus B5 Provide Key Information on Viral Evolution and Survival.
J.Virol., 96, 2022
|
|
7EJV
 
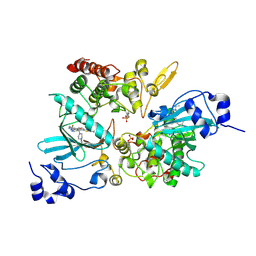 | | The co-crystal structure of DYRK2 with YK-2-69 | | Descriptor: | Dual specificity tyrosine-phosphorylation-regulated kinase 2, [6-[[4-[2-(dimethylamino)-1,3-benzothiazol-6-yl]-5-fluoranyl-pyrimidin-2-yl]amino]pyridin-3-yl]-(4-ethylpiperazin-1-yl)methanone | | Authors: | Li, Z, Xiao, Y, Yuan, K, Kuang, W, Xiuquan, Y, Yang, P. | | Deposit date: | 2021-04-02 | | Release date: | 2022-04-06 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Targeting dual-specificity tyrosine phosphorylation-regulated kinase 2 with a highly selective inhibitor for the treatment of prostate cancer.
Nat Commun, 13, 2022
|
|
7DNO
 
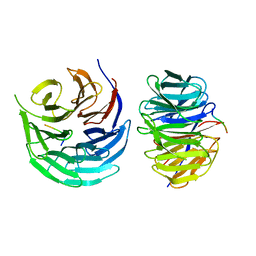 | | Characterization of Peptide Ligands Against WDR5 Isolated Using Phage Display Technique | | Descriptor: | CYS-ARG-THR-LEU-PRO-PHE, WD repeat-containing protein 5 | | Authors: | Cao, J, Cao, D, Xiong, B, Li, Y, Fan, T. | | Deposit date: | 2020-12-10 | | Release date: | 2021-02-10 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.03 Å) | | Cite: | Phage-Display Based Discovery and Characterization of Peptide Ligands against WDR5.
Molecules, 26, 2021
|
|
7XB2
 
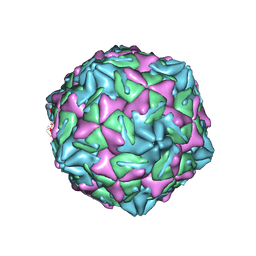 | |
7EY8
 
 | | portal | | Descriptor: | Portal protein | | Authors: | Liu, H.R, Chen, W.Y. | | Deposit date: | 2021-05-30 | | Release date: | 2021-09-22 | | Last modified: | 2024-06-12 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Structural changes in bacteriophage T7 upon receptor-induced genome ejection.
Proc.Natl.Acad.Sci.USA, 118, 2021
|
|
7EY7
 
 | | bacteriophage T7 tail complex | | Descriptor: | Internal virion protein gp14, Tail fiber protein, Tail tubular protein gp11, ... | | Authors: | Liu, H.R, Chen, W.Y. | | Deposit date: | 2021-05-30 | | Release date: | 2021-09-22 | | Last modified: | 2024-06-12 | | Method: | ELECTRON MICROSCOPY (4.3 Å) | | Cite: | Structural changes in bacteriophage T7 upon receptor-induced genome ejection.
Proc.Natl.Acad.Sci.USA, 118, 2021
|
|
