6CZC
 
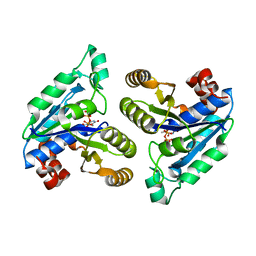 | | Crystal structure of Mycobacterium tuberculosis dethiobiotin synthetase in complex with thymidine triphosphate (TTP) - promiscuous binding mode with disordered nucleoside | | Descriptor: | ATP-dependent dethiobiotin synthetase BioD, MAGNESIUM ION, THYMIDINE-5'-TRIPHOSPHATE | | Authors: | Thompson, A.P, Wegener, K.L, Bruning, J.B, Polyak, S.W. | | Deposit date: | 2018-04-09 | | Release date: | 2018-11-21 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Mycobacterium tuberculosis Dethiobiotin Synthetase Facilitates Nucleoside Triphosphate Promiscuity through Alternate Binding Modes
Acs Catalysis, 8(11), 2018
|
|
6CVK
 
 | |
6CVU
 
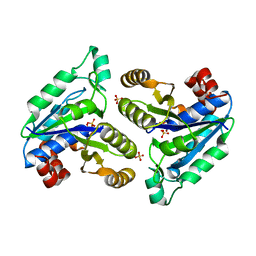 | | Crystal structure of Mycobacterium tuberculosis dethiobiotin synthetase in complex with cytidine | | Descriptor: | 4-AMINO-1-BETA-D-RIBOFURANOSYL-2(1H)-PYRIMIDINONE, ATP-dependent dethiobiotin synthetase BioD, SULFATE ION | | Authors: | Thompson, A.P, Wegener, K.L, Bruning, J.B, Polyak, S.W. | | Deposit date: | 2018-03-28 | | Release date: | 2018-11-21 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.426 Å) | | Cite: | Mycobacterium tuberculosis Dethiobiotin Synthetase Facilitates Nucleoside Triphosphate Promiscuity through Alternate Binding Modes
Acs Catalysis, 8(11), 2018
|
|
6CZE
 
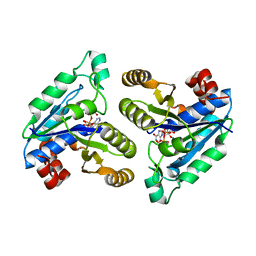 | | Crystal structure of Mycobacterium tuberculosis dethiobiotin synthetase in complex with inosine triphosphate (ITP) - promiscuous binding mode with disordered nucleoside | | Descriptor: | ATP-dependent dethiobiotin synthetase BioD, INOSINE 5'-TRIPHOSPHATE, MAGNESIUM ION | | Authors: | Thompson, A.P, Wegener, K.L, Bruning, J.B, Polyak, S.W. | | Deposit date: | 2018-04-09 | | Release date: | 2018-11-21 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Mycobacterium tuberculosis Dethiobiotin Synthetase Facilitates Nucleoside Triphosphate Promiscuity through Alternate Binding Modes
Acs Catalysis, 8(11), 2018
|
|
3GX2
 
 | |
6CWD
 
 | |
3E33
 
 | | Protein farnesyltransferase complexed with FPP and ethylenediamine scaffold inhibitor 7 | | Descriptor: | FARNESYL DIPHOSPHATE, N-benzyl-N-(2-{(4-cyanophenyl)[(1-methyl-1H-imidazol-5-yl)methyl]amino}ethyl)-2-methylbenzenesulfonamide, Protein farnesyltransferase subunit beta, ... | | Authors: | Hast, M.A, Beese, L.S. | | Deposit date: | 2008-08-06 | | Release date: | 2009-03-10 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structural basis for binding and selectivity of antimalarial and anticancer ethylenediamine inhibitors to protein farnesyltransferase.
Chem.Biol., 16, 2009
|
|
6CIX
 
 | |
6CEJ
 
 | |
3E30
 
 | | Protein farnesyltransferase complexed with FPP and ethylene diamine inhibitor 4 | | Descriptor: | FARNESYL DIPHOSPHATE, Protein farnesyltransferase subunit beta, Protein farnesyltransferase/geranylgeranyltransferase type-1 subunit alpha, ... | | Authors: | Hast, M.A, Beese, L.S. | | Deposit date: | 2008-08-06 | | Release date: | 2009-03-10 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.45 Å) | | Cite: | Structural basis for binding and selectivity of antimalarial and anticancer ethylenediamine inhibitors to protein farnesyltransferase.
Chem.Biol., 16, 2009
|
|
4QV2
 
 | |
6DC0
 
 | |
3FZP
 
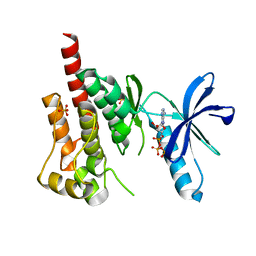 | | Crystal structure of PYK2 complexed with ATPgS | | Descriptor: | PHOSPHOTHIOPHOSPHORIC ACID-ADENYLATE ESTER, Protein tyrosine kinase 2 beta, SULFATE ION | | Authors: | Han, S. | | Deposit date: | 2009-01-26 | | Release date: | 2009-03-31 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structural characterization of proline-rich tyrosine kinase 2 (PYK2) reveals a unique (DFG-out) conformation and enables inhibitor design.
J.Biol.Chem., 284, 2009
|
|
3FZT
 
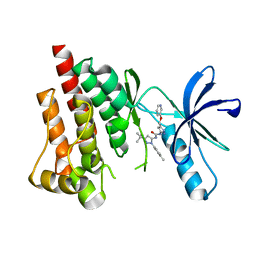 | | Crystal structure of PYK2 complexed with PF-4618433 | | Descriptor: | 1-[5-tert-butyl-2-(4-methylphenyl)-1,2-dihydro-3H-pyrazol-3-ylidene]-3-{3-[(pyridin-3-yloxy)methyl]-1H-pyrazol-5-yl}urea, Protein tyrosine kinase 2 beta | | Authors: | Han, S. | | Deposit date: | 2009-01-26 | | Release date: | 2009-03-31 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Structural characterization of proline-rich tyrosine kinase 2 (PYK2) reveals a unique (DFG-out) conformation and enables inhibitor design.
J.Biol.Chem., 284, 2009
|
|
3WO7
 
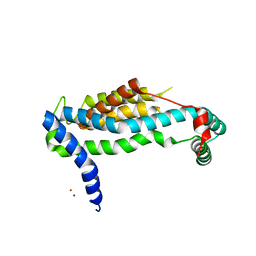 | | Crystal structure of YidC from Bacillus halodurans (form II) | | Descriptor: | COPPER (II) ION, Membrane protein insertase YidC 2 | | Authors: | Kumazaki, K, Tsukazaki, T, Ishitani, R, Nureki, O. | | Deposit date: | 2013-12-20 | | Release date: | 2014-04-23 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (3.201 Å) | | Cite: | Structural basis of Sec-independent membrane protein insertion by YidC.
Nature, 509, 2014
|
|
3WOY
 
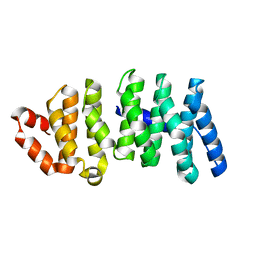 | | Crystal structure of CLASP2 TOG domain (TOG2) | | Descriptor: | CLIP-associating protein 2 | | Authors: | Hayashi, I, Maki, T. | | Deposit date: | 2014-01-06 | | Release date: | 2015-05-27 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | CLASP2 Has Two Distinct TOG Domains That Contribute Differently to Microtubule Dynamics
J. Mol. Biol., 427, 2015
|
|
3GE3
 
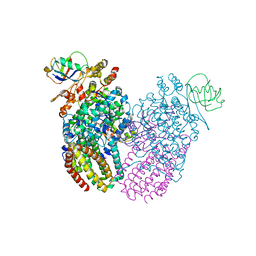 | | Crystal Structure of the reduced Toluene 4-Monooxygenase HD T201A mutant complex | | Descriptor: | ACETATE ION, CHLORIDE ION, FE (III) ION, ... | | Authors: | Elsen, N.L, Bailey, L.J, Fox, B.G. | | Deposit date: | 2009-02-24 | | Release date: | 2009-07-28 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.52 Å) | | Cite: | Role for threonine 201 in the catalytic cycle of the soluble diiron hydroxylase toluene 4-monooxygenase.
Biochemistry, 48, 2009
|
|
3GAB
 
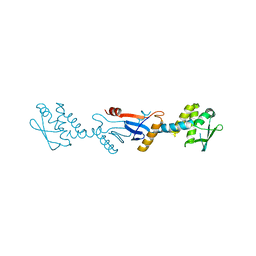 | | C-terminal domain of Bacillus subtilis MutL crystal form I | | Descriptor: | DNA mismatch repair protein mutL | | Authors: | Guarne, A, Pillon, M.C, Lorenowicz, J.J, Mitchell, R.R, Chung, Y.S, Friedhoff, P. | | Deposit date: | 2009-02-17 | | Release date: | 2010-07-21 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structure of the endonuclease domain of MutL: unlicensed to cut.
Mol.Cell, 39, 2010
|
|
3GK2
 
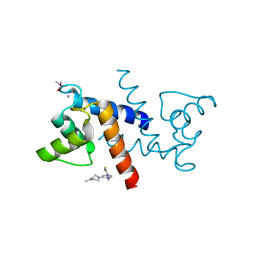 | | X-ray structure of bovine SBi279,Ca(2+)-S100B | | Descriptor: | (Z)-2-[2-(4-methylpiperazin-1-yl)benzyl]diazenecarbothioamide, CACODYLATE ION, CALCIUM ION, ... | | Authors: | Charpentier, T.H, Weber, D.J, Toth, E.A. | | Deposit date: | 2009-03-09 | | Release date: | 2009-06-09 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.984 Å) | | Cite: | Small molecules bound to unique sites in the target protein binding cleft of calcium-bound S100B as characterized by nuclear magnetic resonance and X-ray crystallography.
Biochemistry, 48, 2009
|
|
3GK4
 
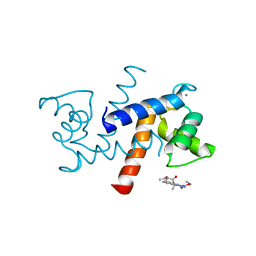 | | X-ray structure of bovine SBi523,Ca(2+)-S100B | | Descriptor: | CALCIUM ION, Protein S100-B, ethyl 5-{[(1R)-1-(ethoxycarbonyl)-2-oxopropyl]sulfanyl}-1,2-dihydro[1,2,3]triazolo[1,5-a]quinazoline-3-carboxylate | | Authors: | Charpentier, T.H, Weber, D.J, Toth, E.A. | | Deposit date: | 2009-03-09 | | Release date: | 2009-06-09 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Small molecules bound to unique sites in the target protein binding cleft of calcium-bound S100B as characterized by nuclear magnetic resonance and X-ray crystallography.
Biochemistry, 48, 2009
|
|
3WBN
 
 | | Crystal structure of MATE in complex with MaL6 | | Descriptor: | (2R)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, MaL6, Putative uncharacterized protein | | Authors: | Tanaka, Y, Ishitani, R, Nureki, O. | | Deposit date: | 2013-05-20 | | Release date: | 2013-06-12 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.45 Å) | | Cite: | Structural basis for the drug extrusion mechanism by a MATE multidrug transporter.
Nature, 496, 2013
|
|
3GK1
 
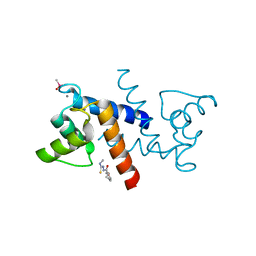 | | X-ray structure of bovine SBi132,Ca(2+)-S100B | | Descriptor: | 2-[(5-hex-1-yn-1-ylfuran-2-yl)carbonyl]-N-methylhydrazinecarbothioamide, CACODYLATE ION, CALCIUM ION, ... | | Authors: | Charpentier, T.H, Weber, D.J, Toth, E.A. | | Deposit date: | 2009-03-09 | | Release date: | 2009-06-09 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Small molecules bound to unique sites in the target protein binding cleft of calcium-bound S100B as characterized by nuclear magnetic resonance and X-ray crystallography.
Biochemistry, 48, 2009
|
|
3WO6
 
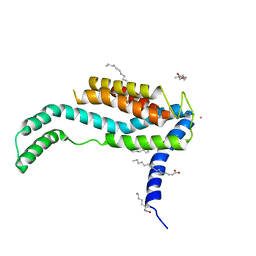 | | Crystal structure of YidC from Bacillus halodurans (form I) | | Descriptor: | (2R)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, CADMIUM ION, Membrane protein insertase YidC 2 | | Authors: | Kumazaki, K, Tsukazaki, T, Ishitani, R, Nureki, O. | | Deposit date: | 2013-12-20 | | Release date: | 2014-04-23 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.403 Å) | | Cite: | Structural basis of Sec-independent membrane protein insertion by YidC.
Nature, 509, 2014
|
|
3W4T
 
 | | Crystal structure of MATE P26A mutant | | Descriptor: | (2R)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, Putative uncharacterized protein | | Authors: | Tanaka, Y, Ishitani, R, Nureki, O. | | Deposit date: | 2013-01-16 | | Release date: | 2013-04-03 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.096 Å) | | Cite: | Structural basis for the drug extrusion mechanism by a MATE multidrug transporter.
Nature, 496, 2013
|
|
3GX5
 
 | |
