7O8J
 
 | | NmHR light state structure at 20 us after photoexcitation determined by serial femtosecond crystallography (with extrapolated, dark and light dataset) | | Descriptor: | (2R)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, CHLORIDE ION, Chloride pumping rhodopsin, ... | | Authors: | Mous, S, Gotthard, G, Ehrenberg, D, Sen, S, James, D, Johnson, P, Weinert, T, Nass, K, Furrer, A, Kekilli, D, Ma, P, Bruenle, S, Casadei, C, Martiel, I, Dworkowski, F, Gashi, D, Skopintsev, P, Wranik, M, Knopp, G, Panepucci, E, Panneels, V, Cirelli, C, Ozerov, D, Schertler, G, Wang, M, Milne, C, Standfuss, J, Schapiro, I, Heberle, J, Nogly, P. | | Deposit date: | 2021-04-15 | | Release date: | 2022-02-09 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Dynamics and mechanism of a light-driven chloride pump.
Science, 375, 2022
|
|
7O8L
 
 | | NmHR dark state structure determined by serial millisecond crystallography | | Descriptor: | (2R)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, CHLORIDE ION, Chloride pumping rhodopsin, ... | | Authors: | Mous, S, Gotthard, G, Ehrenberg, D, Sen, S, James, D, Johnson, P, Weinert, T, Nass, K, Furrer, A, Kekilli, D, Ma, P, Bruenle, S, Casadei, C, Martiel, I, Dworkowski, F, Gashi, D, Skopintsev, P, Wranik, M, Knopp, G, Panepucci, E, Panneels, V, Cirelli, C, Ozerov, D, Schertler, G, Wang, M, Milne, C, Standfuss, J, Schapiro, I, Heberle, J, Nogly, P. | | Deposit date: | 2021-04-15 | | Release date: | 2022-02-09 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Dynamics and mechanism of a light-driven chloride pump.
Science, 375, 2022
|
|
7O8R
 
 | | NmHR light state structure at 27.5 ms (25 - 30 ms) after photoexcitation determined by serial millisecond crystallography | | Descriptor: | (2R)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, CHLORIDE ION, Chloride pumping rhodopsin, ... | | Authors: | Mous, S, Gotthard, G, Ehrenberg, D, Sen, S, James, D, Johnson, P, Weinert, T, Nass, K, Furrer, A, Kekilli, D, Ma, P, Bruenle, S, Casadei, C, Martiel, I, Dworkowski, F, Gashi, D, Skopintsev, P, Wranik, M, Knopp, G, Panepucci, E, Panneels, V, Cirelli, C, Ozerov, D, Schertler, G, Wang, M, Milne, C, Standfuss, J, Schapiro, I, Heberle, J, Nogly, P. | | Deposit date: | 2021-04-15 | | Release date: | 2022-02-09 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Dynamics and mechanism of a light-driven chloride pump.
Science, 375, 2022
|
|
7O8V
 
 | | NmHR light state structure at 55 ms (50 - 60 ms) after photoexcitation determined by serial millisecond crystallography | | Descriptor: | (2R)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, CHLORIDE ION, Chloride pumping rhodopsin, ... | | Authors: | Mous, S, Gotthard, G, Ehrenberg, D, Sen, S, James, D, Johnson, P, Weinert, T, Nass, K, Furrer, A, Kekilli, D, Ma, P, Bruenle, S, Casadei, C, Martiel, I, Dworkowski, F, Gashi, D, Skopintsev, P, Wranik, M, Knopp, G, Panepucci, E, Panneels, V, Cirelli, C, Ozerov, D, Schertler, G, Wang, M, Milne, C, Standfuss, J, Schapiro, I, Heberle, J, Nogly, P. | | Deposit date: | 2021-04-15 | | Release date: | 2022-02-09 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Dynamics and mechanism of a light-driven chloride pump.
Science, 375, 2022
|
|
4YY1
 
 | | The structure of hemagglutinin from a H6N1 influenza virus (A/chicken/Taiwan/A2837/2013) in complex with human receptor analog 6'SLNLN | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, HA1, ... | | Authors: | Wang, F, Qi, J, Bi, Y, Zhang, W, Wang, M, Wang, M, Liu, J, Yan, J, Shi, Y, Gao, G.F. | | Deposit date: | 2015-03-23 | | Release date: | 2016-03-23 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Structure of hemagglutinin from a H6N1 influenza virus (A/chicken/Taiwan/A2837/2013)
To Be Published
|
|
5DQU
 
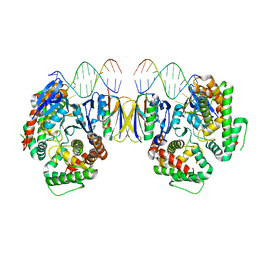 | | Crystal Structure of Cas-DNA-10 complex | | Descriptor: | CRISPR-associated endonuclease Cas1, CRISPR-associated endoribonuclease Cas2, DNA (5'-D(*GP*AP*GP*TP*CP*GP*AP*TP*GP*CP*TP*TP*TP*TP*T)-3'), ... | | Authors: | Wang, J, Li, J, Zhao, H, Sheng, G, Wang, M, Yin, M, Wang, Y. | | Deposit date: | 2015-09-15 | | Release date: | 2015-11-11 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (4.5 Å) | | Cite: | Structural and Mechanistic Basis of PAM-Dependent Spacer Acquisition in CRISPR-Cas Systems.
Cell, 163, 2015
|
|
2WFL
 
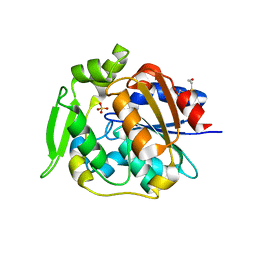 | | Crystal structure of polyneuridine aldehyde esterase | | Descriptor: | POLYNEURIDINE-ALDEHYDE ESTERASE, SULFATE ION | | Authors: | Yang, L, Hill, M, Panjikar, S, Wang, M, Stoeckigt, J. | | Deposit date: | 2009-04-08 | | Release date: | 2009-08-18 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structural Basis and Enzymatic Mechanism of the Biosynthesis of C9- from C10-Monoterpenoid Indole Alkaloids.
Angew.Chem.Int.Ed.Engl., 48, 2009
|
|
5DQT
 
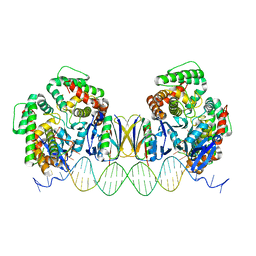 | | Crystal Structure of Cas-DNA-22 complex | | Descriptor: | CRISPR-associated endonuclease Cas1, CRISPR-associated endoribonuclease Cas2, DNA (33-MER), ... | | Authors: | Wang, J, Li, J, Zhao, H, Sheng, G, Wang, M, Yin, M, Wang, Y. | | Deposit date: | 2015-09-15 | | Release date: | 2015-11-11 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Structural and Mechanistic Basis of PAM-Dependent Spacer Acquisition in CRISPR-Cas Systems.
Cell, 163, 2015
|
|
5DQZ
 
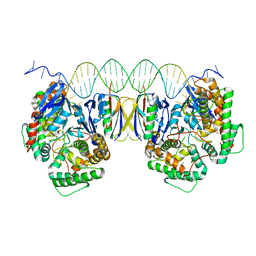 | | Crystal Structure of Cas-DNA-PAM complex | | Descriptor: | CRISPR-associated endonuclease Cas1, CRISPR-associated endoribonuclease Cas2, DNA (36-MER), ... | | Authors: | Wang, J, Li, J, Zhao, H, Sheng, G, Wang, M, Yin, M, Wang, Y. | | Deposit date: | 2015-09-15 | | Release date: | 2015-11-11 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Structural and Mechanistic Basis of PAM-Dependent Spacer Acquisition in CRISPR-Cas Systems.
Cell, 163, 2015
|
|
6FMV
 
 | | IMISX-EP of Hg-BacA soaking SIRAS | | Descriptor: | (2R)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, CITRATE ANION, ... | | Authors: | Huang, C.-Y, Olieric, V, Howe, N, Warshamanage, R, Weinert, T, Panepucci, E, Vogeley, L, Basu, S, Diederichs, K, Caffrey, M, Wang, M. | | Deposit date: | 2018-02-02 | | Release date: | 2018-09-05 | | Last modified: | 2024-06-19 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | In situ serial crystallography for rapid de novo membrane protein structure determination.
Commun Biol, 1, 2018
|
|
6FMX
 
 | | IMISX-EP of W-PgpB | | Descriptor: | (2R)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, Phosphatidylglycerophosphatase B, TUNGSTATE(VI)ION | | Authors: | Huang, C.-Y, Olieric, V, Howe, N, Warshamanage, R, Weinert, T, Panepucci, E, Vogeley, L, Basu, S, Diederichs, K, Caffrey, M, Wang, M. | | Deposit date: | 2018-02-02 | | Release date: | 2018-09-05 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.79 Å) | | Cite: | In situ serial crystallography for rapid de novo membrane protein structure determination.
Commun Biol, 1, 2018
|
|
2WFM
 
 | | Crystal structure of polyneuridine aldehyde esterase mutant (H244A) | | Descriptor: | POLYNEURIDINE ALDEHYDE ESTERASE | | Authors: | Yang, L, Hill, M, Panjikar, S, Wang, M, Stoeckigt, J. | | Deposit date: | 2009-04-08 | | Release date: | 2010-04-21 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structural Basis and Enzymatic Mechanism of the Biosynthesis of C9- from C10-Monoterpenoid Indole Alkaloids.
Angew.Chem.Int.Ed.Engl., 48, 2009
|
|
6FMS
 
 | | IMISX-EP of Se-LspA | | Descriptor: | (2R)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, Globomycin, Lipoprotein signal peptidase | | Authors: | Huang, C.-Y, Olieric, V, Howe, N, Warshamanage, R, Weinert, T, Panepucci, E, Vogeley, L, Basu, S, Diederichs, K, Caffrey, M, Wang, M. | | Deposit date: | 2018-02-02 | | Release date: | 2018-08-29 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | In situ serial crystallography for rapid de novo membrane protein structure determination.
Commun Biol, 1, 2018
|
|
6FMR
 
 | | IMISX-EP of Se-PepTSt | | Descriptor: | (2S)-2,3-DIHYDROXYPROPYL(7Z)-PENTADEC-7-ENOATE, Di-or tripeptide:H+ symporter, PHOSPHATE ION | | Authors: | Huang, C.-Y, Olieric, V, Howe, N, Warshamanage, R, Weinert, T, Panepucci, E, Vogeley, L, Basu, S, Diederichs, K, Caffrey, M, Wang, M. | | Deposit date: | 2018-02-02 | | Release date: | 2018-08-29 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | In situ serial crystallography for rapid de novo membrane protein structure determination.
Commun Biol, 1, 2018
|
|
6FMW
 
 | | IMISX-EP of Hg-BacA cocrystallization | | Descriptor: | (2R)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, MERCURY (II) ION, ... | | Authors: | Huang, C.-Y, Olieric, V, Howe, N, Warshamanage, R, Weinert, T, Panepucci, E, Vogeley, L, Basu, S, Diederichs, K, Caffrey, M, Wang, M. | | Deposit date: | 2018-02-02 | | Release date: | 2018-09-05 | | Last modified: | 2024-06-19 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | In situ serial crystallography for rapid de novo membrane protein structure determination.
Commun Biol, 1, 2018
|
|
6FMT
 
 | | IMISX-EP of Hg-BacA Soaking SAD | | Descriptor: | (2R)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, MERCURY (II) ION, ... | | Authors: | Huang, C.-Y, Olieric, V, Howe, N, Warshamanage, R, Weinert, T, Panepucci, E, Vogeley, L, Basu, S, Diederichs, K, Caffrey, M, Wang, M. | | Deposit date: | 2018-02-02 | | Release date: | 2018-09-05 | | Last modified: | 2024-06-19 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | In situ serial crystallography for rapid de novo membrane protein structure determination.
Commun Biol, 1, 2018
|
|
6FMY
 
 | | IMISX-EP of S-PepTSt | | Descriptor: | (2S)-2,3-DIHYDROXYPROPYL(7Z)-PENTADEC-7-ENOATE, 3,6,9,12,15,18,21,24-OCTAOXAHEXACOSAN-1-OL, Di-or tripeptide:H+ symporter, ... | | Authors: | Huang, C.-Y, Olieric, V, Howe, N, Warshamanage, R, Weinert, T, Panepucci, E, Vogeley, L, Basu, S, Diederichs, K, Caffrey, M, Wang, M. | | Deposit date: | 2018-02-02 | | Release date: | 2018-09-05 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | In situ serial crystallography for rapid de novo membrane protein structure determination.
Commun Biol, 1, 2018
|
|
5DLJ
 
 | | Crystal Structure of Cas-DNA-N1 complex | | Descriptor: | 39-mer DNA N1-F, 39-mer DNA N1-R, CRISPR-associated endonuclease Cas1, ... | | Authors: | Wang, J, Li, J, Zhao, H, Sheng, G, Wang, M, Yin, M, Wang, Y. | | Deposit date: | 2015-09-05 | | Release date: | 2015-11-11 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structural and Mechanistic Basis of PAM-Dependent Spacer Acquisition in CRISPR-Cas Systems.
Cell, 163, 2015
|
|
4YY9
 
 | | The structure of hemagglutinin from a H6N1 influenza virus (A/Taiwan/2/2013) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, HA1, HA2, ... | | Authors: | Wang, F, Qi, J, Bi, Y, Zhang, W, Wang, M, Wang, M, Liu, J, Yan, J, Shi, Y, Gao, G.F. | | Deposit date: | 2015-03-23 | | Release date: | 2016-04-13 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.601 Å) | | Cite: | Structure of hemagglutinin from a H6N1 influenza virus (A/Taiwan/2/2013) at 2.6 Angstroms resolution
To Be Published
|
|
4YY7
 
 | | The structure of hemagglutinin from a H6N1 influenza virus (A/chicken/Taiwan/A2837/2013) in complex with avian receptor analog 3'SLNLN | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, HA1, ... | | Authors: | Wang, F, Qi, J, Bi, Y, Zhang, W, Wang, M, Wang, M, Liu, J, Yan, J, Shi, Y, Gao, G.F. | | Deposit date: | 2015-03-23 | | Release date: | 2016-03-23 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.99 Å) | | Cite: | Structure of hemagglutinin from a H6N1 influenza virus (A/chicken/Taiwan/A2837/2013)
To Be Published
|
|
4YY0
 
 | | The structure of hemagglutinin from a H6N1 influenza virus (A/chicken/Taiwan/A2837/2013) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, HA1, HA2 | | Authors: | Wang, F, Qi, J, Bi, Y, Zhang, W, Wang, M, Wang, M, Liu, J, Yan, J, Shi, Y, Gao, G.F. | | Deposit date: | 2015-03-23 | | Release date: | 2016-03-23 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.59 Å) | | Cite: | Structure of hemagglutinin from a H6N1 influenza virus (A/chicken/Taiwan/A2837/2013)
To Be Published
|
|
4YYA
 
 | | The structure of hemagglutinin from a H6N1 influenza virus (A/Taiwan/2/2013) in complex with avian receptor analog 3'SLNLN | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, HA1, HA2, ... | | Authors: | Wang, F, Qi, J, Bi, Y, Zhang, W, Wang, M, Wang, M, Liu, J, Yan, J, Shi, Y, Gao, G.F. | | Deposit date: | 2015-03-23 | | Release date: | 2016-04-13 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.595 Å) | | Cite: | Structure of hemagglutinin from a H6N1 influenza virus (A/chicken/Taiwan/A2837/2013)
To Be Published
|
|
4YYB
 
 | | The structure of hemagglutinin from a H6N1 influenza virus (A/Taiwan/2/2013) in complex with human receptor analog 6'SLNLN | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, HA1, HA2, ... | | Authors: | Wang, F, Qi, J, Bi, Y, Zhang, W, Wang, M, Wang, M, Liu, J, Yan, J, Shi, Y, Gao, G.F. | | Deposit date: | 2015-03-23 | | Release date: | 2016-04-13 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.609 Å) | | Cite: | Structure of hemagglutinin from a H6N1 influenza virus (A/Taiwan/2/2013) at 2.6 Angstroms resolution
To Be Published
|
|
4ZWJ
 
 | | Crystal structure of rhodopsin bound to arrestin by femtosecond X-ray laser | | Descriptor: | Chimera protein of human Rhodopsin, mouse S-arrestin, and T4 Endolysin | | Authors: | Kang, Y, Zhou, X.E, Gao, X, He, Y, Liu, W, Ishchenko, A, Barty, A, White, T.A, Yefanov, O, Han, G.W, Xu, Q, de Waal, P.W, Ke, J, Tan, M.H.E, Zhang, C, Moeller, A, West, G.M, Pascal, B, Eps, N.V, Caro, L.N, Vishnivetskiy, S.A, Lee, R.J, Suino-Powell, K.M, Gu, X, Pal, K, Ma, J, Zhi, X, Boutet, S, Williams, G.J, Messerschmidt, M, Gati, C, Zatsepin, N.A, Wang, D, James, D, Basu, S, Roy-Chowdhury, S, Conrad, C, Coe, J, Liu, H, Lisova, S, Kupitz, C, Grotjohann, I, Fromme, R, Jiang, Y, Tan, M, Yang, H, Li, J, Wang, M, Zheng, Z, Li, D, Howe, N, Zhao, Y, Standfuss, J, Diederichs, K, Dong, Y, Potter, C.S, Carragher, B, Caffrey, M, Jiang, H, Chapman, H.N, Spence, J.C.H, Fromme, P, Weierstall, U, Ernst, O.P, Katritch, V, Gurevich, V.V, Griffin, P.R, Hubbell, W.L, Stevens, R.C, Cherezov, V, Melcher, K, Xu, H.E, GPCR Network (GPCR) | | Deposit date: | 2015-05-19 | | Release date: | 2015-07-29 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (3.302 Å) | | Cite: | Crystal structure of rhodopsin bound to arrestin by femtosecond X-ray laser.
Nature, 523, 2015
|
|
6S0L
 
 | | Structure of the A2A adenosine receptor determined at SwissFEL using native-SAD at 4.57 keV from all available diffraction patterns | | Descriptor: | (2R)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, (2S)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, 4-{2-[(7-amino-2-furan-2-yl[1,2,4]triazolo[1,5-a][1,3,5]triazin-5-yl)amino]ethyl}phenol, ... | | Authors: | Nass, K, Cheng, R, Vera, L, Mozzanica, A, Redford, S, Ozerov, D, Basu, S, James, D, Knopp, G, Cirelli, C, Martiel, I, Casadei, C, Weinert, T, Nogly, P, Skopintsev, P, Usov, I, Leonarski, F, Geng, T, Rappas, M, Dore, A.S, Cooke, R, Nasrollahi Shirazi, S, Dworkowski, F, Sharpe, M, Olieric, N, Steinmetz, M.O, Schertler, G, Abela, R, Patthey, L, Schmitt, B, Hennig, M, Standfuss, J, Wang, M, Milne, J.C. | | Deposit date: | 2019-06-17 | | Release date: | 2020-07-15 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.65 Å) | | Cite: | Advances in long-wavelength native phasing at X-ray free-electron lasers.
Iucrj, 7, 2020
|
|
