1Y2I
 
 | | Crystal Structure of MCSG Target APC27401 from Shigella flexneri | | Descriptor: | Hypothetical Protein S0862 | | Authors: | Brunzelle, J.S, Minasov, G, Yang, X, Shuvalova, L, Collart, F.R, Anderson, W.F, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2004-11-22 | | Release date: | 2005-01-04 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal Structure of MCSG Target APC27401 from Shigella flexneri
To be Published
|
|
2AUA
 
 | | Structure of BC2332: A Protein of Unknown Function from Bacillus cereus | | Descriptor: | hypothetical protein | | Authors: | Brunzelle, J.S, Minasov, G, Shuvalova, L, Collart, F.R, Joachimiak, A, Anderson, W.F, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2005-08-27 | | Release date: | 2005-10-11 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Crystal structure of BC2332: A hypothetical protein from Bacillus cereus
To be Published
|
|
1X7F
 
 | | Crystal structure of an uncharacterized B. cereus protein | | Descriptor: | outer surface protein | | Authors: | Minasov, G, Shuvalova, L, Brunzelle, J.S, Collart, F.R, Anderson, W.F, MCSG, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2004-08-13 | | Release date: | 2004-08-24 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal structure of an uncharacterized B. cereus protein
To be Published
|
|
2AP1
 
 | | Crystal structure of the putative regulatory protein | | Descriptor: | SODIUM ION, ZINC ION, putative regulator protein | | Authors: | Brunzelle, J.S, Minasov, G, Shuvalova, L, Collart, F.R, Joachimiak, A, Anderson, W.F, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2005-08-15 | | Release date: | 2005-09-27 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal structure of the putative regulatory protein
To be Published
|
|
2BAS
 
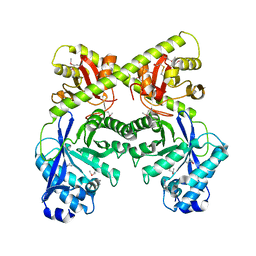 | | Crystal Structure of the Bacillus subtilis YkuI Protein, with an EAL Domain. | | Descriptor: | BETA-MERCAPTOETHANOL, YkuI protein | | Authors: | Minasov, G, Brunzelle, J.S, Shuvalova, L, Miller, D.J, Collart, F.R, Joachimiak, A, Anderson, W.F, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2005-10-14 | | Release date: | 2005-11-29 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (2.61 Å) | | Cite: | Crystal structures of YkuI and its complex with second messenger cyclic Di-GMP suggest catalytic mechanism of phosphodiester bond cleavage by EAL domains.
J.Biol.Chem., 284, 2009
|
|
3HJB
 
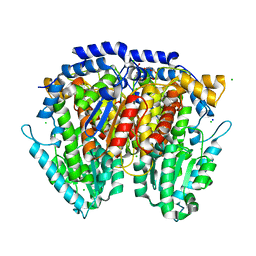 | | 1.5 Angstrom Crystal Structure of Glucose-6-phosphate Isomerase from Vibrio cholerae. | | Descriptor: | CALCIUM ION, CHLORIDE ION, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Minasov, G, Halavaty, A, Shuvalova, L, Dubrovska, I, Winsor, J, Papazisi, L, Anderson, W.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2009-05-21 | | Release date: | 2009-06-16 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | 1.5 Angstrom Crystal Structure of Glucose-6-phosphate Isomerase from Vibrio cholerae.
To be Published
|
|
3IJ3
 
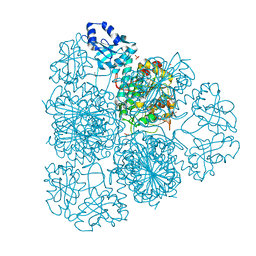 | | 1.8 Angstrom Resolution Crystal Structure of Cytosol Aminopeptidase from Coxiella burnetii | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, Cytosol aminopeptidase, ... | | Authors: | Minasov, G, Halavaty, A, Shuvalova, L, Dubrovska, I, Winsor, J, Papazisi, L, Anderson, W.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2009-08-03 | | Release date: | 2009-08-11 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | 1.8 Angstrom Resolution Crystal Structure of Cytosol Aminopeptidase from Coxiella burnetii
TO BE PUBLISHED
|
|
3IJ5
 
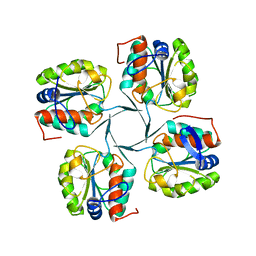 | | 1.95 Angstrom Resolution Crystal Structure of 3-deoxy-D-manno-octulosonate 8-phosphate phosphatase from Yersinia pestis | | Descriptor: | 3-deoxy-D-manno-octulosonate 8-phosphate phosphatase, CHLORIDE ION | | Authors: | Minasov, G, Halavaty, A, Shuvalova, L, Dubrovska, I, Winsor, J, Papazisi, L, Anderson, W.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2009-08-03 | | Release date: | 2009-08-11 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | 1.95 Angstrom Resolution Crystal Structure of 3-deoxy-D-manno-octulosonate 8-phosphate phosphatase from Yersinia pestis
TO BE PUBLISHED
|
|
3JZE
 
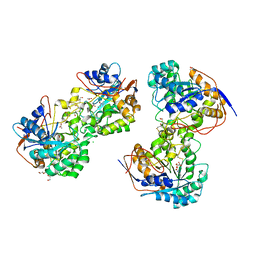 | | 1.8 Angstrom resolution crystal structure of dihydroorotase (pyrC) from Salmonella enterica subsp. enterica serovar Typhimurium str. LT2 | | Descriptor: | ACETIC ACID, BETA-MERCAPTOETHANOL, CHLORIDE ION, ... | | Authors: | Minasov, G, Halavaty, A, Shuvalova, L, Dubrovska, I, Winsor, J, Papazisi, L, Anderson, W.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2009-09-23 | | Release date: | 2009-09-29 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | 1.8 Angstrom Resolution Crystal Structure of Dihydroorotase (pyrC) from Salmonella enterica subsp. enterica serovar Typhimurium str. LT2.
TO BE PUBLISHED
|
|
3HL3
 
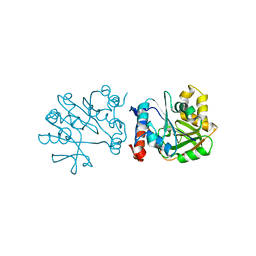 | | 2.76 Angstrom Crystal Structure of a Putative Glucose-1-Phosphate Thymidylyltransferase from Bacillus anthracis in Complex with a Sucrose. | | Descriptor: | Glucose-1-phosphate thymidylyltransferase, beta-D-fructofuranose-(2-1)-alpha-D-glucopyranose | | Authors: | Minasov, G, Shuvalova, L, Halavaty, A, Dubrovska, I, Winsor, J, Papazisi, L, Anderson, W.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2009-05-26 | | Release date: | 2009-06-09 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.76 Å) | | Cite: | 2.76 Angstrom Crystal Structure of a Putative Glucose-1-Phosphate Thymidylyltransferase from Bacillus anthracis in Complex with a Sucrose.
TO BE PUBLISHED
|
|
3KB8
 
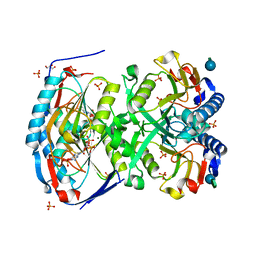 | | 2.09 Angstrom resolution structure of a hypoxanthine-guanine phosphoribosyltransferase (hpt-1) from Bacillus anthracis str. 'Ames Ancestor' in complex with GMP | | Descriptor: | GLYCEROL, GUANOSINE-5'-MONOPHOSPHATE, Hypoxanthine phosphoribosyltransferase, ... | | Authors: | Halavaty, A.S, Minasov, G, Shuvalova, L, Dubrovska, I, Winsor, J, Papazisi, L, Anderson, W.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2009-10-20 | | Release date: | 2009-10-27 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.09 Å) | | Cite: | 2.09 Angstrom resolution structure of a hypoxanthine-guanine phosphoribosyltransferase (hpt-1) from Bacillus anthracis str. 'Ames Ancestor' in complex with GMP
To be Published
|
|
3HVU
 
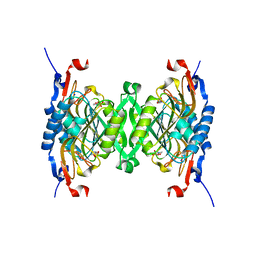 | | 1.95 Angstrom Crystal Structure of Complex of Hypoxanthine-Guanine Phosphoribosyltransferase from Bacillus anthracis with 2-(N-morpholino)ethanesulfonic acid (MES) | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, Hypoxanthine phosphoribosyltransferase, SODIUM ION | | Authors: | Minasov, G, Halavaty, A, Shuvalova, L, Dubrovska, I, Winsor, J, Papazisi, L, Anderson, W.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2009-06-16 | | Release date: | 2009-06-23 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | 1.95 Angstrom Crystal Structure of Complex of Hypoxanthine-Guanine Phosphoribosyltransferase from Bacillus anthracis with 2-(N-morpholino)ethanesulfonic acid (MES)
TO BE PUBLISHED
|
|
7L6R
 
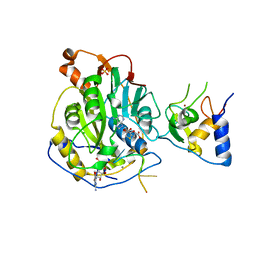 | | Crystal Structure of SARS-CoV-2 Nsp16/10 Heterodimer in Complex with (m7GpppA2m)pUpUpApApA (Cap-1), S-Adenosyl-L-homocysteine (SAH) and Manganese (Mn). | | Descriptor: | 2'-O-methyltransferase, CHLORIDE ION, MANGANESE (II) ION, ... | | Authors: | Minasov, G, Shuvalova, L, Rosas-Lemus, M, Kiryukhina, O, Brunzelle, J.S, Satchell, K.J.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2020-12-23 | | Release date: | 2021-01-06 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.98 Å) | | Cite: | Mn 2+ coordinates Cap-0-RNA to align substrates for efficient 2'- O -methyl transfer by SARS-CoV-2 nsp16.
Sci.Signal., 14, 2021
|
|
7L6T
 
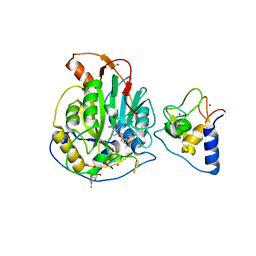 | | Crystal Structure of SARS-CoV-2 Nsp16/10 Heterodimer in Complex with (m7GpppA2m)pUpUpApApA (Cap-1), S-Adenosyl-L-homocysteine (SAH) and two Magnesium (Mg) ions. | | Descriptor: | 2'-O-methyltransferase, CHLORIDE ION, FORMIC ACID, ... | | Authors: | Minasov, G, Shuvalova, L, Rosas-Lemus, M, Kiryukhina, O, Brunzelle, J.S, Satchell, K.J.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2020-12-23 | | Release date: | 2021-01-06 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.78 Å) | | Cite: | Mn 2+ coordinates Cap-0-RNA to align substrates for efficient 2'- O -methyl transfer by SARS-CoV-2 nsp16.
Sci.Signal., 14, 2021
|
|
7MQ5
 
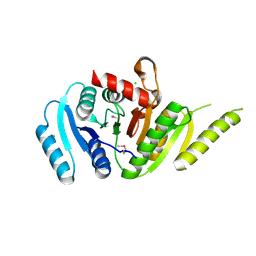 | | Crystal Structure of Putative Universal Stress Protein from Pseudomonas aeruginosa UCBPP-PA14 | | Descriptor: | CHLORIDE ION, Universal stress protein | | Authors: | Minasov, G, Shuvalova, L, Kiryukhina, O, Dubrovska, I, Satchell, K.J.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2021-05-05 | | Release date: | 2021-05-19 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.25 Å) | | Cite: | Crystal Structure of Putative Universal Stress Protein from Pseudomonas aeruginosa UCBPP-PA14
To Be Published
|
|
7L75
 
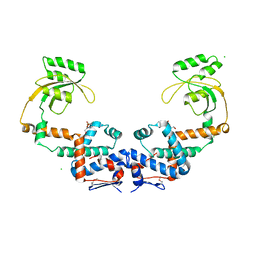 | | Crystal Structure of Peptidylprolyl Isomerase PrsA from Streptococcus mutans. | | Descriptor: | 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, CHLORIDE ION, Foldase protein PrsA | | Authors: | Minasov, G, Shuvalova, L, Kiryukhina, O, Wawrzak, Z, Satchell, K.J.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2020-12-25 | | Release date: | 2021-12-01 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (3.15 Å) | | Cite: | Crystal Structure of Peptidylprolyl Isomerase PrsA from Streptococcus mutans.
To Be Published
|
|
7L6L
 
 | | Crystal Structure of the DNA-binding Transcriptional Repressor DeoR from Escherichia coli str. K-12 | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, Deoxyribose operon repressor, ... | | Authors: | Minasov, G, Shuvalova, L, Kiryukhina, O, Dubrovska, I, Wiersum, G, Satchell, K.J.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2020-12-23 | | Release date: | 2021-12-01 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Crystal Structure of the DNA-binding Transcriptional Repressor DeoR from Escherichia coli str. K-12.
To Be Published
|
|
7L6J
 
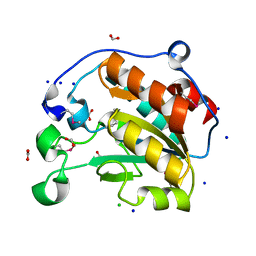 | | Crystal Structure of the Putative Hydrolase from Stenotrophomonas maltophilia | | Descriptor: | CHLORIDE ION, FORMIC ACID, Putative hydrolase, ... | | Authors: | Minasov, G, Shuvalova, L, Kiryukhina, O, Dubrovska, I, Wiersum, G, Satchell, K.J.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2020-12-23 | | Release date: | 2021-12-01 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.78 Å) | | Cite: | Crystal Structure of the Putative Hydrolase from Stenotrophomonas maltophilia
To Be Published
|
|
7L6Y
 
 | |
7L6Z
 
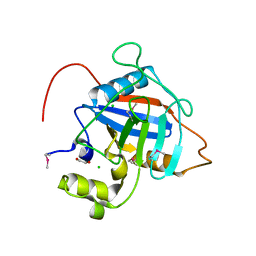 | | Crystal Structure of Peptidyl-Prolyl Cis-Trans Isomerasefrom (PpiB) Streptococcus pneumoniae R6 | | Descriptor: | 1,2-ETHANEDIOL, 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, CHLORIDE ION, ... | | Authors: | Minasov, G, Shuvalova, L, Kiryukhina, O, Dubrovska, I, Satchell, K.J.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2020-12-24 | | Release date: | 2021-12-01 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.88 Å) | | Cite: | Crystal Structure of Peptidyl-Prolyl Cis-Trans Isomerasefrom (PpiB) Streptococcus pneumoniae R6
To Be Published
|
|
7L71
 
 | |
7L5T
 
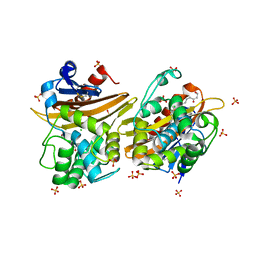 | | Crystal Structure of the Oxacillin-hydrolyzing Class D Extended-spectrum Beta-lactamase OXA-14 from Pseudomonas aeruginosa in Complex with Covalently Bound Clavulanic Acid | | Descriptor: | (2E)-3-[(4-hydroxy-2-oxobutyl)amino]prop-2-enal, Beta-lactamase, CHLORIDE ION, ... | | Authors: | Minasov, G, Shuvalova, L, Rosas-Lemus, M, Brunzelle, J.S, Satchell, K.J.F, Center for Structural Genomics of Infectious Diseases (CSGID), Center for Membrane Proteins of Infectious Diseases (MPID) | | Deposit date: | 2020-12-22 | | Release date: | 2021-12-29 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.88 Å) | | Cite: | Crystal Structure of the Oxacillin-hydrolyzing Class D Extended-spectrum Beta-lactamase OXA-14 from Pseudomonas aeruginosa in Complex with Covalently Bound Clavulanic Acid
To Be Published
|
|
6AWA
 
 | | 1.83 Angstrom Resolution Crystal Structure of Dihydrolipoyl Dehydrogenase from Pseudomonas putida in Complex with FAD and Adenosine-5'-monophosphate. | | Descriptor: | ADENOSINE MONOPHOSPHATE, Dihydrolipoyl dehydrogenase, FLAVIN-ADENINE DINUCLEOTIDE, ... | | Authors: | Minasov, G, Shuvalova, L, Kiryukhina, O, Dubrovska, I, Grimshaw, S, Kwon, K, Anderson, W.F, Satchell, K.J.F, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2017-09-05 | | Release date: | 2017-10-04 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.83 Å) | | Cite: | 1.83 Angstrom Resolution Crystal Structure of Dihydrolipoyl Dehydrogenase from Pseudomonas putida in Complex with FAD and Adenosine-5'-monophosphate.
To Be Published
|
|
6AZI
 
 | | 1.75 Angstrom Resolution Crystal Structure of D-alanyl-D-alanine Endopeptidase from Enterobacter cloacae in Complex with Covalently Bound Boronic Acid | | Descriptor: | BORATE ION, D-alanyl-D-alanine endopeptidase | | Authors: | Minasov, G, Wawrzak, Z, Shuvalova, L, Kiryukhina, O, Dubrovska, I, Grimshaw, S, Kwon, K, Anderson, W.F, Satchell, K.J.F, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2017-09-11 | | Release date: | 2017-10-04 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | 1.75 Angstrom Resolution Crystal Structure of D-alanyl-D-alanine Endopeptidase from Enterobacter cloacae in Complex with Covalently Bound Boronic Acid.
To be Published
|
|
6B8D
 
 | | 1.78 Angstrom Resolution Crystal Structure of N-terminal Fragment (residues 1-405) of Elongation Factor G from Haemophilus influenzae | | Descriptor: | CHLORIDE ION, Elongation factor G | | Authors: | Minasov, G, Shuvalova, L, Dubrovska, I, Kiryukhina, O, Grimshaw, S, Kwon, K, Anderson, W.F, Satchell, K.J.F, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2017-10-06 | | Release date: | 2017-10-18 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.78 Å) | | Cite: | 1.78 Angstrom Resolution Crystal Structure of N-terminal Fragment (residues 1-405) of Elongation Factor G from Haemophilus influenzae.
To Be Published
|
|
