5ZL9
 
 | |
5XCY
 
 | | Structure of the cellobiohydrolase Cel6A from Phanerochaete chrysosporium at 1.2 angstrom | | Descriptor: | Glucanase | | Authors: | Tachioka, M, Nakamura, A, Ishida, T, Igarashi, K, Samejima, M. | | Deposit date: | 2017-03-24 | | Release date: | 2017-07-26 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (1.199 Å) | | Cite: | Crystal structure of a family 6 cellobiohydrolase from the basidiomycete Phanerochaete chrysosporium
Acta Crystallogr F Struct Biol Commun, 73, 2017
|
|
6IY9
 
 | | Crystal structure of aminoglycoside 7"-phoshotransferase-Ia (APH(7")-Ia/HYG) from Streptomyces hygroscopicus complexed with hygromycin B | | Descriptor: | 2-[N-CYCLOHEXYLAMINO]ETHANE SULFONIC ACID, CITRATE ANION, HYGROMYCIN B VARIANT, ... | | Authors: | Takenoya, M, Shimamura, T, Yamanaka, R, Adachi, Y, Ito, S, Sasaki, Y, Nakamura, A, Yajima, S. | | Deposit date: | 2018-12-14 | | Release date: | 2019-09-11 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structural basis for the substrate recognition of aminoglycoside 7''-phosphotransferase-Ia from Streptomyces hygroscopicus.
Acta Crystallogr.,Sect.F, 75, 2019
|
|
4TWL
 
 | | Crystal structure of dioscorin complexed with ascorbate | | Descriptor: | ASCORBIC ACID, Dioscorin 5, SULFATE ION | | Authors: | Xue, Y.L, Miyakawa, T, Nakamura, A, Tanokura, M. | | Deposit date: | 2014-07-01 | | Release date: | 2015-04-01 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.11 Å) | | Cite: | Yam Tuber Storage Protein Reduces Plant Oxidants Using the Coupled Reactions as Carbonic Anhydrase and Dehydroascorbate Reductase
Mol Plant, 8, 2015
|
|
7Y8U
 
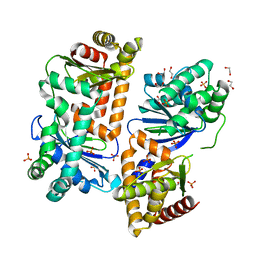 | | Crystal structure of AlbEF homolog from Quasibacillus thermotolerans | | Descriptor: | 1,2-ETHANEDIOL, AlbE homolog, AlbF homolog, ... | | Authors: | Ishida, K, Nakamura, A, Kojima, S. | | Deposit date: | 2022-06-24 | | Release date: | 2022-10-12 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystal structure of the AlbEF complex involved in subtilosin A biosynthesis.
Structure, 30, 2022
|
|
7Y8V
 
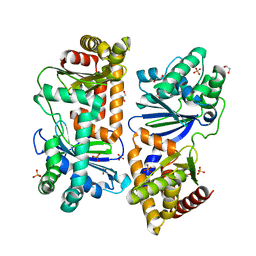 | | Crystal structure of AlbEF homolog mutant (AlbF-H54A/H58A) from Quasibacillus thermotolerans | | Descriptor: | 1,2-ETHANEDIOL, AlbE homolog, AlbF homolog H54A/H58A mutant, ... | | Authors: | Ishida, K, Nakamura, A, Kojima, S. | | Deposit date: | 2022-06-24 | | Release date: | 2022-10-12 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal structure of the AlbEF complex involved in subtilosin A biosynthesis.
Structure, 30, 2022
|
|
7Y8X
 
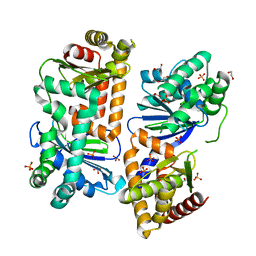 | | Crystal structure of AlbEF homolog from Quasibacillus thermotolerans in complex with Ni(II) | | Descriptor: | 1,2-ETHANEDIOL, AlbE homolog, AlbF homolog, ... | | Authors: | Ishida, K, Nakamura, A, Kojima, S. | | Deposit date: | 2022-06-24 | | Release date: | 2022-10-12 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Crystal structure of the AlbEF complex involved in subtilosin A biosynthesis.
Structure, 30, 2022
|
|
2KTT
 
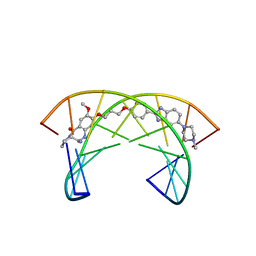 | | Solution Structure of a Covalently Bound Pyrrolo[2,1-c][1,4]benzodiazepine-Benzimidazole Hybrid to a 10mer DNA Duplex | | Descriptor: | (11aS)-7-methoxy-8-(3-{4-[6-(4-methylpiperazin-1-yl)-1H-benzimidazol-2-yl]phenoxy}propoxy)-1,2,3,10,11,11a-hexahydro-5H-pyrrolo[2,1-c][1,4]benzodiazepin-5-one, 5'-D(*AP*AP*CP*AP*AP*TP*TP*GP*TP*T)-3' | | Authors: | Rettig, M, Weingarth, M, Langel, W, Kamal, A, Kumar, P.P, Weisz, K. | | Deposit date: | 2010-02-08 | | Release date: | 2010-03-09 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Solution structure of a covalently bound pyrrolo[2,1-c][1,4]benzodiazepine-benzimidazole hybrid to a 10mer DNA duplex.
Biochemistry, 48, 2009
|
|
2KY7
 
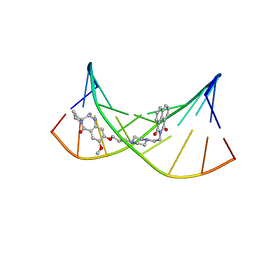 | | NMR Structural Studies on the Covalent DNA Binding of a Pyrrolobenzodiazepine-Naphthalimide Conjugate | | Descriptor: | 2-{2-[4-(3-{[(11aS)-7-methoxy-5-oxo-2,3,5,10,11,11a-hexahydro-1H-pyrrolo[2,1-c][1,4]benzodiazepin-8-yl]oxy}propyl)piperazin-1-yl]ethyl}-1H-benzo[de]isoquinoline-1,3(2H)-dione, 5'-D(*AP*AP*CP*AP*AP*TP*TP*GP*TP*T)-3' | | Authors: | Rettig, M, Langel, W, Kamal, A, Weisz, K. | | Deposit date: | 2010-05-17 | | Release date: | 2010-06-02 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | NMR structural studies on the covalent DNA binding of a pyrrolobenzodiazepine-naphthalimide conjugate
Org.Biomol.Chem., 8, 2010
|
|
4GA6
 
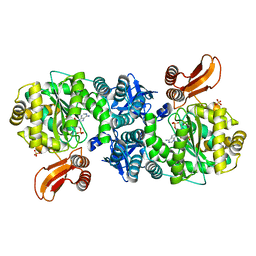 | | Crystal structure of AMP phosphorylase C-terminal deletion mutant in complex with substrates | | Descriptor: | ADENOSINE MONOPHOSPHATE, Putative thymidine phosphorylase, SULFATE ION | | Authors: | Nishitani, Y, Aono, R, Nakamura, A, Sato, T, Atomi, H, Imanaka, T, Miki, K. | | Deposit date: | 2012-07-25 | | Release date: | 2013-05-15 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.21 Å) | | Cite: | Structure analysis of archaeal AMP phosphorylase reveals two unique modes of dimerization
J.Mol.Biol., 425, 2013
|
|
1PL7
 
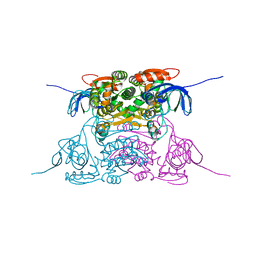 | | Human Sorbitol Dehydrogenase (apo) | | Descriptor: | Sorbitol dehydrogenase, ZINC ION | | Authors: | Pauly, T.A, Ekstrom, J.L, Beebe, D.A, Chrunyk, B, Cunningham, D, Griffor, M, Kamath, A, Lee, S.E, Madura, R, Mcguire, D, Subashi, T, Wasilko, D, Watts, P, Mylari, B.L, Oates, P.J, Adams, P.D, Rath, V.L. | | Deposit date: | 2003-06-07 | | Release date: | 2004-02-17 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | X-ray crystallographic and kinetic studies of human sorbitol dehydrogenase.
Structure, 11, 2003
|
|
1PL8
 
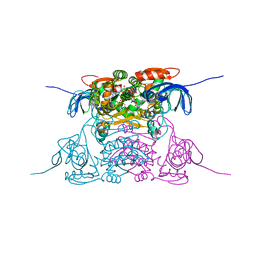 | | human SDH/NAD+ complex | | Descriptor: | NICOTINAMIDE-ADENINE-DINUCLEOTIDE, ZINC ION, human sorbitol dehydrogenase | | Authors: | Pauly, T.A, Ekstrom, J.L, Beebe, D.A, Chrunyk, B, Cunningham, D, Griffor, M, Kamath, A, Lee, S.E, Madura, R, Mcguire, D, Subashi, T, Wasilko, D, Watts, P, Mylari, B.L, Oates, P.J, Adams, P.D, Rath, V.L. | | Deposit date: | 2003-06-07 | | Release date: | 2004-02-17 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | X-ray crystallographic and kinetic studies of human sorbitol dehydrogenase.
Structure, 11, 2003
|
|
1PL6
 
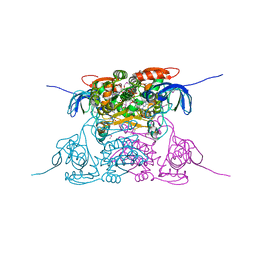 | | Human SDH/NADH/inhibitor complex | | Descriptor: | 4-[2-(HYDROXYMETHYL)PYRIMIDIN-4-YL]-N,N-DIMETHYLPIPERAZINE-1-SULFONAMIDE, NICOTINAMIDE-ADENINE-DINUCLEOTIDE, Sorbitol dehydrogenase, ... | | Authors: | Pauly, T.A, Ekstrom, J.L, Beebe, D.A, Chrunyk, B, Cunningham, D, Griffor, M, Kamath, A, Lee, S.E, Madura, R, Mcguire, D, Subashi, T, Wasilko, D, Watts, P, Mylari, B.L, Oates, P.J, Adams, P.D, Rath, V.L. | | Deposit date: | 2003-06-07 | | Release date: | 2004-02-17 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | X-ray crystallographic and kinetic studies of human sorbitol dehydrogenase.
Structure, 11, 2003
|
|
4GA5
 
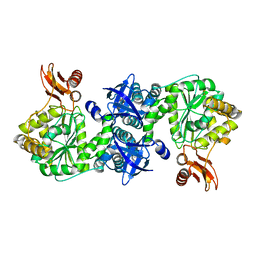 | | Crystal structure of AMP phosphorylase C-terminal deletion mutant in the apo-form | | Descriptor: | Putative thymidine phosphorylase | | Authors: | Nishitani, Y, Aono, R, Nakamura, A, Sato, T, Atomi, H, Imanaka, T, Miki, K. | | Deposit date: | 2012-07-25 | | Release date: | 2013-05-15 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (3.25 Å) | | Cite: | Structure analysis of archaeal AMP phosphorylase reveals two unique modes of dimerization
J.Mol.Biol., 425, 2013
|
|
4GA4
 
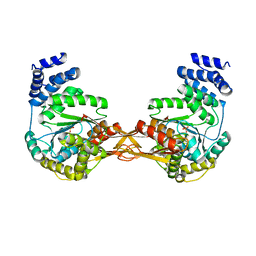 | | Crystal structure of AMP phosphorylase N-terminal deletion mutant | | Descriptor: | PHOSPHATE ION, Putative thymidine phosphorylase | | Authors: | Nishitani, Y, Aono, R, Nakamura, A, Sato, T, Atomi, H, Imanaka, T, Miki, K. | | Deposit date: | 2012-07-25 | | Release date: | 2013-05-15 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (3.51 Å) | | Cite: | Structure analysis of archaeal AMP phosphorylase reveals two unique modes of dimerization
J.Mol.Biol., 425, 2013
|
|
1WUN
 
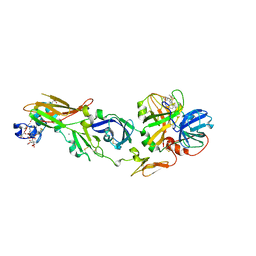 | | Human Factor Viia-Tissue Factor Complexed with ethylsulfonamide-D-Trp-Gln-p-aminobenzamidine | | Descriptor: | CALCIUM ION, Coagulation factor VII, N-(ETHYLSULFONYL)TRYPTOPHYL-N~1~-{4-[AMINO(IMINO)METHYL]BENZYL}GLUTAMAMIDE, ... | | Authors: | Kadono, S, Sakamoto, A, Kikuchi, Y, Oh-eda, M, Yabuta, N, Yoshihashi, K, Kitazawa, T, Suzuki, T, Koga, T, Hattori, K, Shiraishi, T, Haramura, M, Kodama, H, Ono, Y, Esaki, T, Sato, H, Watanabe, Y, Itoh, S, Ohta, M, Kozono, T. | | Deposit date: | 2004-12-08 | | Release date: | 2005-12-08 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structure-based design of P3 moieties in the peptide mimetic factor VIIa inhibitor
Biochem.Biophys.Res.Commun., 327, 2005
|
|
1WV7
 
 | | Human Factor Viia-Tissue Factor Complexed with ethylsulfonamide-D-5-propoxy-Trp-Gln-p-aminobenzamidine | | Descriptor: | CALCIUM ION, Coagulation factor VII, N-(ETHYLSULFONYL)-5-PROPOXY-L-TRYPTOPHYL-N~1~-{4-[AMINO(IMINO)METHYL]BENZYL}-L-GLUTAMAMIDE, ... | | Authors: | Kadono, S, Sakamoto, A, Kikuchi, Y, Oh-eda, M, Yabuta, N, Yoshihashi, K, Kitazawa, T, Suzuki, T, Koga, T, Hattori, K, Shiraishi, T, Haramura, M, Kodama, H, Ono, Y, Esaki, T, Sato, H, Watanabe, Y, Itoh, S, Ohta, M, Kozono, T. | | Deposit date: | 2004-12-11 | | Release date: | 2005-12-11 | | Last modified: | 2025-03-26 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Structure-based design of P3 moieties in the peptide mimetic factor VIIa inhibitor
Biochem.Biophys.Res.Commun., 327, 2005
|
|
1WSS
 
 | | Human Factor Viia-Tissue Factor in Complex with peptide-mimetic inhibitor that has two charged groups in P2 and P4 | | Descriptor: | CALCIUM ION, Coagulation factor VII, N-[(3-CARBOXYBENZYL)SULFONYL]ISOLEUCYL-N~1~-{4-[AMINO(IMINO)METHYL]BENZYL}-5-IMINOORNITHINAMIDE, ... | | Authors: | Kadono, S, Sakamoto, A, Kikuchi, Y, Oh-Eda, M, Yabuta, N, Koga, T, Hattori, K, Shiraishi, T, Haramura, M, Kodama, H, Ono, Y, Esaki, T, Sato, H, Watanabe, Y, Itoh, S, Ohta, M, Kozono, T. | | Deposit date: | 2004-11-10 | | Release date: | 2005-11-10 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structure of human factor VIIa/tissue factor in complex with a peptide-mimetic inhibitor: high selectivity against thrombin by introducing two charged groups in P2 and P4.
Acta Crystallogr.,Sect.F, 61, 2005
|
|
1YOO
 
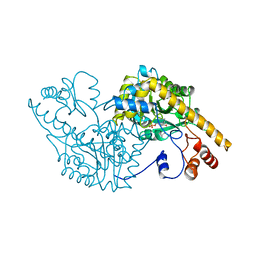 | | ASPARTATE AMINOTRANSFERASE MUTANT ATB17 WITH ISOVALERIC ACID | | Descriptor: | ASPARTATE AMINOTRANSFERASE, ISOVALERIC ACID, PYRIDOXAL-5'-PHOSPHATE | | Authors: | Oue, S, Okamoto, A, Yano, T, Kagamiyama, H. | | Deposit date: | 1998-06-26 | | Release date: | 1999-02-02 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Redesigning the substrate specificity of an enzyme by cumulative effects of the mutations of non-active site residues.
J.Biol.Chem., 274, 1999
|
|
2E0I
 
 | | Crystal structure of archaeal photolyase from Sulfolobus tokodaii with two FAD molecules: Implication of a novel light-harvesting cofactor | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, 432aa long hypothetical deoxyribodipyrimidine photolyase, FLAVIN-ADENINE DINUCLEOTIDE | | Authors: | Fujihashi, M, Numoto, N, Kobayashi, Y, Mizushima, A, Tsujimura, M, Nakamura, A, Kawarabayashi, Y, Miki, K. | | Deposit date: | 2006-10-10 | | Release date: | 2006-11-28 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Crystal Structure of Archaeal Photolyase from Sulfolobus tokodaii with Two FAD Molecules: Implication of a Novel Light-harvesting Cofactor
J.Mol.Biol., 365, 2007
|
|
2ATE
 
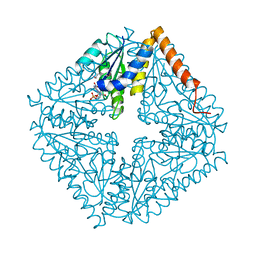 | | Structure of the complex of PurE with NitroAIR | | Descriptor: | ((2R,3S,4R,5R)-5-(5-AMINO-4-NITRO-1H-IMIDAZOL-1-YL)-3,4-DIHYDROXYTETRAHYDROFURAN-2-YL)METHYL DIHYDROGEN PHOSPHATE, Phosphoribosylaminoimidazole carboxylase catalytic subunit | | Authors: | Kappock, T.J, Mathews, I.I, Zaugg, J.B, Peng, P, Hoskins, A.A, Okamoto, A, Ealick, S.E, Stubbe, J. | | Deposit date: | 2005-08-24 | | Release date: | 2006-08-29 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | N5-CAIR mutase: role of a CO2 binding site and substrate movement in catalysis.
Biochemistry, 46, 2007
|
|
2D57
 
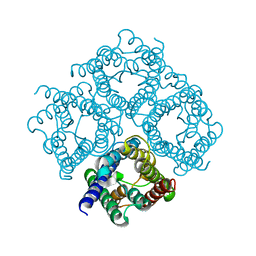 | | Double layered 2D crystal structure of AQUAPORIN-4 (AQP4M23) at 3.2 a resolution by electron crystallography | | Descriptor: | Aquaporin-4 | | Authors: | Hiroaki, Y, Tani, K, Kamegawa, A, Gyobu, N, Nishikawa, K, Suzuki, H, Walz, T, Sasaki, S, Mitsuoka, K, Kimura, K, Mizoguchi, A, Fujiyoshi, Y. | | Deposit date: | 2005-10-29 | | Release date: | 2006-01-31 | | Last modified: | 2023-11-08 | | Method: | ELECTRON CRYSTALLOGRAPHY (3.2 Å) | | Cite: | Implications of the Aquaporin-4 Structure on Array Formation and Cell Adhesion
J.Mol.Biol., 355, 2005
|
|
1UI0
 
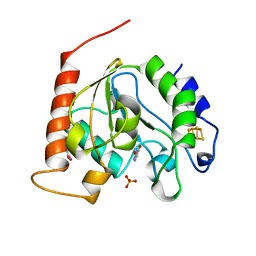 | | Crystal Structure Of Uracil-DNA Glycosylase From Thermus Thermophilus HB8 | | Descriptor: | IRON/SULFUR CLUSTER, SULFATE ION, URACIL, ... | | Authors: | Hoseki, J, Okamoto, A, Masui, R, Shibata, T, Inoue, Y, Yokoyama, S, Kuramitsu, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2003-07-14 | | Release date: | 2003-10-14 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Crystal Structure of a Family 4 Uracil-DNA Glycosylase from Thermus thermophilus HB8
J.Mol.Biol., 333, 2003
|
|
1UI1
 
 | | Crystal Structure Of Uracil-DNA Glycosylase From Thermus Thermophilus HB8 | | Descriptor: | IRON/SULFUR CLUSTER, Uracil-DNA Glycosylase | | Authors: | Hoseki, J, Okamoto, A, Masui, R, Shibata, T, Inoue, Y, Yokoyama, S, Kuramitsu, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2003-07-14 | | Release date: | 2003-10-14 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Crystal Structure of a Family 4 Uracil-DNA Glycosylase from Thermus thermophilus HB8
J.Mol.Biol., 333, 2003
|
|
3VKR
 
 | | Assimilatory nitrite reductase (Nii3) - NO2 complex from tobbaco leaf analysed with high X-ray dose | | Descriptor: | CHLORIDE ION, IRON/SULFUR CLUSTER, NITRITE ION, ... | | Authors: | Nakano, S, Takahashi, M, Sakamoto, A, Morikawa, H, Katayanagi, K. | | Deposit date: | 2011-11-20 | | Release date: | 2012-04-25 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | The reductive reaction mechanism of tobacco nitrite reductase derived from a combination of crystal structures and ultraviolet-visible microspectroscopy
Proteins, 80, 2012
|
|
