2HRW
 
 | | Crystal Structure of Phosphonopyruvate Hydrolase | | Descriptor: | CHLORIDE ION, Phosphonopyruvate hydrolase, SODIUM ION | | Authors: | Chen, C.C.H, Herzberg, O. | | Deposit date: | 2006-07-20 | | Release date: | 2006-10-03 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structure and Kinetics of Phosphonopyruvate Hydrolase from Voriovorax sp. Pal2: New Insight into the Divergence of Catalysis within the PEP Mutase/Isocitrate Lyase Superfamily
Biochemistry, 45, 2006
|
|
1MWQ
 
 | | Structure of HI0828, a Hypothetical Protein from Haemophilus influenzae with a Putative Active-Site Phosphohistidine | | Descriptor: | CACODYLATE ION, CHLORIDE ION, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Willis, M.A, Krajewski, W, Chalamasetty, V.R, Reddy, P, Howard, A, Herzberg, O, Structure 2 Function Project (S2F) | | Deposit date: | 2002-09-30 | | Release date: | 2003-11-25 | | Last modified: | 2019-07-24 | | Method: | X-RAY DIFFRACTION (0.99 Å) | | Cite: | Structure of YciI from Haemophilus influenzae (HI0828) reveals a ferredoxin-like alpha/beta-fold with a histidine/aspartate centered catalytic site
Proteins, 59, 2005
|
|
1YLI
 
 | |
1PSU
 
 | | Structure of the E. coli PaaI protein from the phyenylacetic acid degradation operon | | Descriptor: | Phenylacetic acid degradation protein PaaI | | Authors: | Kniewel, R, Buglino, J, Solorzano, V, Wu, J, Lima, C.D, Burley, S.K, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2003-06-21 | | Release date: | 2003-07-08 | | Last modified: | 2021-02-03 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structure, function, and mechanism of the phenylacetate pathway hot dog-fold thioesterase PaaI.
J.Biol.Chem., 281, 2006
|
|
1NZY
 
 | |
1PYM
 
 | |
3GAY
 
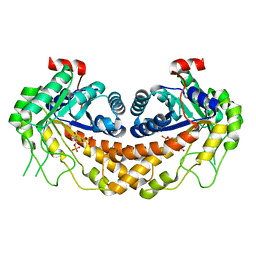 | | Structure of Giardia fructose-1,6-biphosphate aldolase in complex with tagatose-1,6-biphosphate | | Descriptor: | 1,6-di-O-phosphono-D-tagatose, Fructose-bisphosphate aldolase, ZINC ION | | Authors: | Galkin, A, Herzberg, O. | | Deposit date: | 2009-02-18 | | Release date: | 2009-03-31 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structural insights into the substrate binding and stereoselectivity of giardia fructose-1,6-bisphosphate aldolase.
Biochemistry, 48, 2009
|
|
3GB6
 
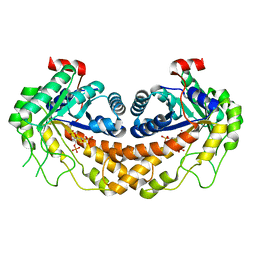 | | Structure of Giardia fructose-1,6-biphosphate aldolase D83A mutant in complex with fructose-1,6-bisphosphate | | Descriptor: | 1,6-di-O-phosphono-D-fructose, Fructose-bisphosphate aldolase, ZINC ION | | Authors: | Galkin, A, Herzberg, O. | | Deposit date: | 2009-02-18 | | Release date: | 2009-03-31 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural insights into the substrate binding and stereoselectivity of giardia fructose-1,6-bisphosphate aldolase.
Biochemistry, 48, 2009
|
|
3GAK
 
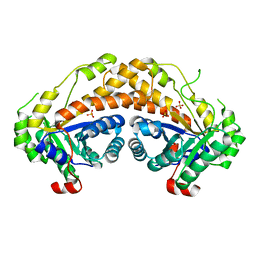 | | Structure of Giardia fructose-1,6-biphosphate aldolase | | Descriptor: | Fructose-bisphosphate aldolase, SULFATE ION, ZINC ION | | Authors: | Galkin, A, Herzberg, O. | | Deposit date: | 2009-02-17 | | Release date: | 2009-03-31 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Structural insights into the substrate binding and stereoselectivity of giardia fructose-1,6-bisphosphate aldolase.
Biochemistry, 48, 2009
|
|
3FA3
 
 | | Crystal structure of 2,3-dimethylmalate lyase, a PEP mutase/isocitrate lyase superfamily member, trigonal crystal form | | Descriptor: | 2,2-difluoro-3,3-dihydroxybutanedioic acid, 2,3-dimethylmalate lyase, GLYCEROL, ... | | Authors: | Narayanan, B.C, Herzberg, O. | | Deposit date: | 2008-11-14 | | Release date: | 2009-01-27 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structure and function of 2,3-dimethylmalate lyase, a PEP mutase/isocitrate lyase superfamily member.
J.Mol.Biol., 386, 2009
|
|
3GRF
 
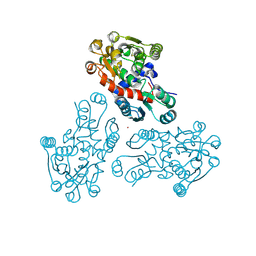 | |
3FA4
 
 | | Crystal structure of 2,3-dimethylmalate lyase, a PEP mutase/isocitrate lyase superfamily member, triclinic crystal form | | Descriptor: | 2,3-dimethylmalate lyase, MAGNESIUM ION | | Authors: | Narayanan, B.C, Herzberg, O. | | Deposit date: | 2008-11-14 | | Release date: | 2009-01-27 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.18 Å) | | Cite: | Structure and function of 2,3-dimethylmalate lyase, a PEP mutase/isocitrate lyase superfamily member.
J.Mol.Biol., 386, 2009
|
|
4OLC
 
 | |
4HGN
 
 | |
4HGO
 
 | |
4HGR
 
 | |
4HGQ
 
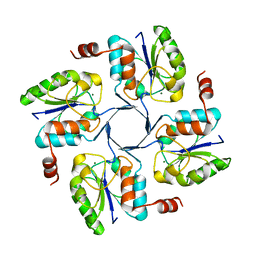 | |
1YPP
 
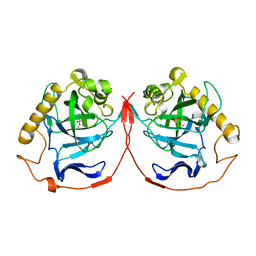 | | ACID ANHYDRIDE HYDROLASE | | Descriptor: | INORGANIC PYROPHOSPHATASE, MANGANESE (II) ION, PHOSPHATE ION | | Authors: | Harutyunyan, E.H, Kuranova, I.P, Lamzin, V.S, Dauter, Z, Wilson, K.S. | | Deposit date: | 1996-05-29 | | Release date: | 1996-12-07 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | X-ray structure of yeast inorganic pyrophosphatase complexed with manganese and phosphate.
Eur.J.Biochem., 239, 1996
|
|
4JZ9
 
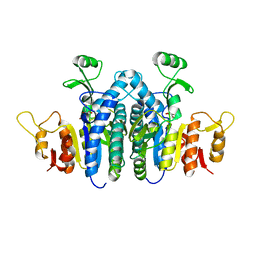 | |
4JZ8
 
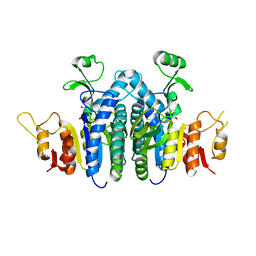 | |
4JZ7
 
 | | Carbamate kinase from Giardia lamblia bound to AMP-PNP | | Descriptor: | Carbamate kinase, PHOSPHOAMINOPHOSPHONIC ACID-ADENYLATE ESTER | | Authors: | Lim, K, Herzberg, O. | | Deposit date: | 2013-04-02 | | Release date: | 2013-06-05 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Crystal Structures of Carbamate Kinase from Giardia lamblia Bound with Citric Acid and AMP-PNP.
Plos One, 8, 2013
|
|
