7ZZP
 
 | | Structure of HDAC2 complexed with an inhibitory ligand | | Descriptor: | (2S)-2-HYDROXYPROPANOIC ACID, 1,2-ETHANEDIOL, 2-[N-CYCLOHEXYLAMINO]ETHANE SULFONIC ACID, ... | | Authors: | Cleasby, A, Tisi, D. | | Deposit date: | 2022-05-25 | | Release date: | 2022-09-21 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.52 Å) | | Cite: | Fragment-Based Discovery of a Novel, Brain Penetrant, Orally Active HDAC2 Inhibitor.
Acs Med.Chem.Lett., 13, 2022
|
|
7ZZR
 
 | | HDAC2 in complex with inhibitory ligand | | Descriptor: | 1,2-ETHANEDIOL, 2-[N-CYCLOHEXYLAMINO]ETHANE SULFONIC ACID, CALCIUM ION, ... | | Authors: | Cleasby, A, Tisi, D. | | Deposit date: | 2022-05-26 | | Release date: | 2022-09-21 | | Last modified: | 2024-06-19 | | Method: | X-RAY DIFFRACTION (2.168 Å) | | Cite: | Fragment-Based Discovery of a Novel, Brain Penetrant, Orally Active HDAC2 Inhibitor.
Acs Med.Chem.Lett., 13, 2022
|
|
7ZZW
 
 | | Ligand binding to HDAC2 | | Descriptor: | 1,2-ETHANEDIOL, 2-(cyclohexylazaniumyl)ethanesulfonate, CALCIUM ION, ... | | Authors: | Cleasby, A, Tisi, D. | | Deposit date: | 2022-05-26 | | Release date: | 2022-09-21 | | Last modified: | 2024-06-19 | | Method: | X-RAY DIFFRACTION (1.73 Å) | | Cite: | Fragment-Based Discovery of a Novel, Brain Penetrant, Orally Active HDAC2 Inhibitor.
Acs Med.Chem.Lett., 13, 2022
|
|
8A0B
 
 | | Inhibitor binding to HDAC2 | | Descriptor: | 1,2-ETHANEDIOL, 1,3-dihydroisoindol-2-yl-[(2R,4S)-4-phenylpyrrolidin-1-ium-2-yl]methanone, 2-[N-CYCLOHEXYLAMINO]ETHANE SULFONIC ACID, ... | | Authors: | Cleasby, A, Tisi, D. | | Deposit date: | 2022-05-27 | | Release date: | 2022-09-21 | | Last modified: | 2024-06-19 | | Method: | X-RAY DIFFRACTION (1.746 Å) | | Cite: | Fragment-Based Discovery of a Novel, Brain Penetrant, Orally Active HDAC2 Inhibitor.
Acs Med.Chem.Lett., 13, 2022
|
|
7ZZS
 
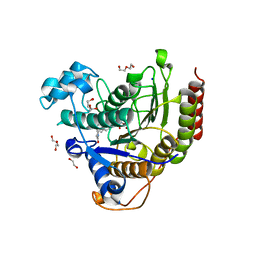 | | HDAC2 complexed with an inhibitory ligand | | Descriptor: | (5~{S})-5-(4-chlorophenyl)pyrrolidin-2-one, 1,2-ETHANEDIOL, 2-(cyclohexylazaniumyl)ethanesulfonate, ... | | Authors: | Cleasby, A, Tisi, D. | | Deposit date: | 2022-05-26 | | Release date: | 2022-09-21 | | Last modified: | 2024-06-19 | | Method: | X-RAY DIFFRACTION (1.88 Å) | | Cite: | Fragment-Based Discovery of a Novel, Brain Penetrant, Orally Active HDAC2 Inhibitor.
Acs Med.Chem.Lett., 13, 2022
|
|
7ZZU
 
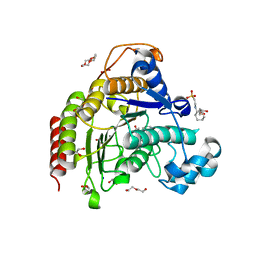 | | Inhibitory Ligand binding to HDAC2 | | Descriptor: | 1,2-ETHANEDIOL, 2-[4-[(2~{R},4~{S})-4-phenylpyrrolidin-2-yl]carbonylpiperazin-1-yl]pyridine-3-carbonitrile, 2-[N-CYCLOHEXYLAMINO]ETHANE SULFONIC ACID, ... | | Authors: | Cleasby, A, Tisi, D. | | Deposit date: | 2022-05-26 | | Release date: | 2022-09-21 | | Last modified: | 2024-06-19 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Fragment-Based Discovery of a Novel, Brain Penetrant, Orally Active HDAC2 Inhibitor.
Acs Med.Chem.Lett., 13, 2022
|
|
6KYW
 
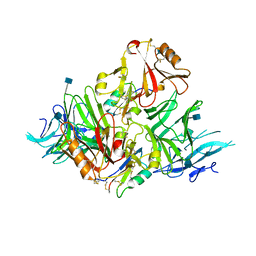 | | S8-mSRK-S8-SP11 complex | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Receptor protein kinase SRK8, S locus protein 11 | | Authors: | Murase, K, Hakoshima, T, Mori, T. | | Deposit date: | 2019-09-20 | | Release date: | 2020-09-16 | | Last modified: | 2020-10-21 | | Method: | X-RAY DIFFRACTION (2.60112739 Å) | | Cite: | Mechanism of self/nonself-discrimination in Brassica self-incompatibility.
Nat Commun, 11, 2020
|
|
2Z83
 
 | |
2ZYE
 
 | | Structure of HIV-1 Protease in Complex with Potent Inhibitor KNI-272 Determined by Neutron Crystallography | | Descriptor: | (4R)-N-tert-butyl-3-[(2S,3S)-2-hydroxy-3-({N-[(isoquinolin-5-yloxy)acetyl]-S-methyl-L-cysteinyl}amino)-4-phenylbutanoyl]-1,3-thiazolidine-4-carboxamide, protease | | Authors: | Adachi, M, Ohhara, T, Tamada, T, Okazaki, N, Kuroki, R. | | Deposit date: | 2009-01-20 | | Release date: | 2009-03-24 | | Last modified: | 2024-05-29 | | Method: | NEUTRON DIFFRACTION (1.9 Å) | | Cite: | Structure of HIV-1 protease in complex with potent inhibitor KNI-272 determined by high-resolution X-ray and neutron crystallography.
Proc.Natl.Acad.Sci.USA, 2009
|
|
3A72
 
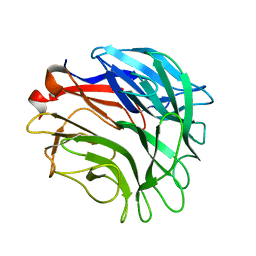 | |
3A71
 
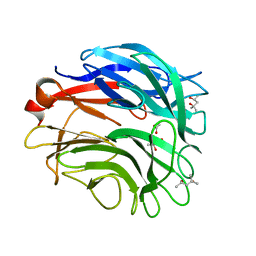 | |
2E2S
 
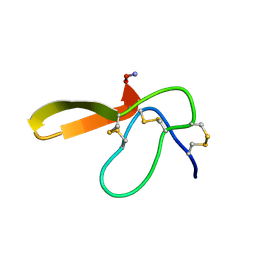 | |
1WXY
 
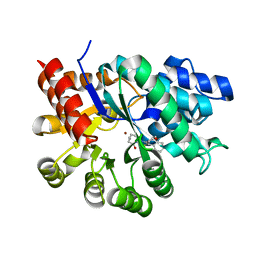 | |
1V49
 
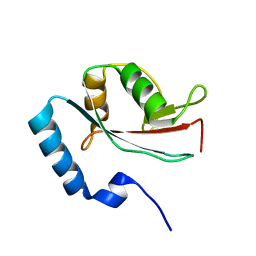 | | Solution structure of microtubule-associated protein light chain-3 | | Descriptor: | Microtubule-associated proteins 1A/1B light chain 3B | | Authors: | Kouno, T, Mizuguchi, M, Tanida, I, Ueno, T, Kominami, E, Kawano, K. | | Deposit date: | 2003-11-11 | | Release date: | 2004-12-28 | | Last modified: | 2023-12-27 | | Method: | SOLUTION NMR | | Cite: | Solution structure of microtubule-associated protein light chain 3 and identification of its functional subdomains.
J.Biol.Chem., 280, 2005
|
|
1VFL
 
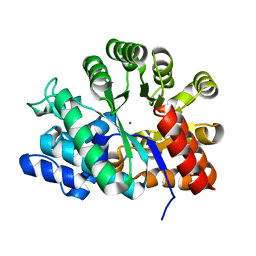 | | Adenosine deaminase | | Descriptor: | Adenosine deaminase, ZINC ION | | Authors: | Kinoshita, T. | | Deposit date: | 2004-04-16 | | Release date: | 2005-08-16 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structural Basis of Compound Recognition by Adenosine Deaminase
Biochemistry, 44, 2005
|
|
2E2F
 
 | |
5B6F
 
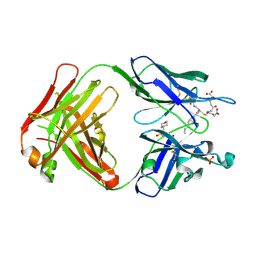 | | Crystal structure of the Fab fragment of an anti-Leukotriene C4 monoclonal antibody complexed with LTC4 | | Descriptor: | (5~{S},6~{R},7~{E},9~{E},11~{Z},14~{Z})-6-[(2~{R})-2-[[(4~{S})-4-azanyl-5-oxidanyl-5-oxidanylidene-pentanoyl]amino]-3-(2-hydroxy-2-oxoethylamino)-3-oxidanylidene-propyl]sulfanyl-5-oxidanyl-icosa-7,9,11,14-tetraenoic acid, 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, SULFATE ION, ... | | Authors: | Sugahara, M, Ago, H, Saino, H, Miyano, M. | | Deposit date: | 2016-05-27 | | Release date: | 2017-05-31 | | Last modified: | 2019-07-24 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystal structure of the Fab fragment of an anti-Leukotriene C4 monoclonal antibody complexed with LTC4
To Be Published
|
|
6L93
 
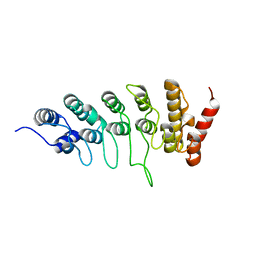 | |
1JDA
 
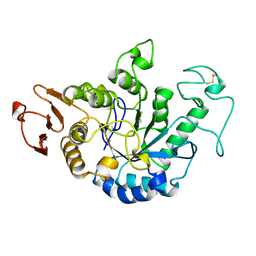 | | MALTOTETRAOSE-FORMING EXO-AMYLASE | | Descriptor: | 1,4-ALPHA MALTOTETRAHYDROLASE, CALCIUM ION | | Authors: | Yoshioka, Y, Hasegawa, K, Matsuura, Y, Katsube, Y, Kubota, M. | | Deposit date: | 1997-06-16 | | Release date: | 1997-10-15 | | Last modified: | 2021-11-03 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal structures of a mutant maltotetraose-forming exo-amylase cocrystallized with maltopentaose.
J.Mol.Biol., 271, 1997
|
|
2RDV
 
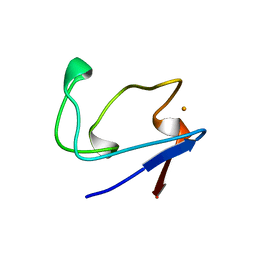 | |
1RDV
 
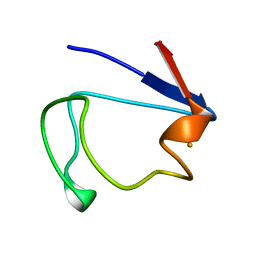 | |
1SRD
 
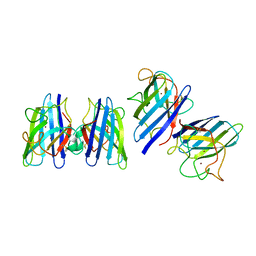 | | Three-dimensional structure of CU,ZN-superoxide dismutase from spinach at 2.0 Angstroms resolution | | Descriptor: | COPPER (II) ION, COPPER,ZINC SUPEROXIDE DISMUTASE, ZINC ION | | Authors: | Kitagawa, Y, Katsube, Y. | | Deposit date: | 1993-04-15 | | Release date: | 1994-01-31 | | Last modified: | 2024-06-05 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Three-dimensional structure of Cu,Zn-superoxide dismutase from spinach at 2.0 A resolution.
J.Biochem.(Tokyo), 109, 1991
|
|
1QI3
 
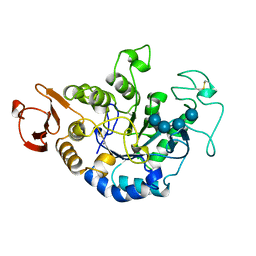 | | MUTANT (D193N) MALTOTETRAOSE-FORMING EXO-AMYLASE IN COMPLEX WITH MALTOTETRAOSE | | Descriptor: | CALCIUM ION, PROTEIN (EXO-MALTOTETRAOHYDROLASE), alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose | | Authors: | Hasegawa, K, Kubota, M, Matsuura, Y. | | Deposit date: | 1999-06-01 | | Release date: | 1999-11-24 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Roles of catalytic residues in alpha-amylases as evidenced by the structures of the product-complexed mutants of a maltotetraose-forming amylase.
Protein Eng., 12, 1999
|
|
1QPK
 
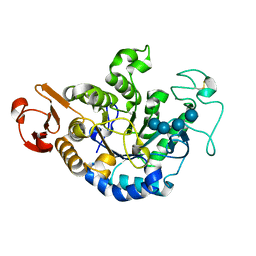 | | MUTANT (D193G) MALTOTETRAOSE-FORMING EXO-AMYLASE IN COMPLEX WITH MALTOTETRAOSE | | Descriptor: | CALCIUM ION, PROTEIN (MALTOTETRAOSE-FORMING AMYLASE), alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose | | Authors: | Yoshioka, Y, Hasegawa, K, Matsuura, Y, Katsube, Y, Kubota, M. | | Deposit date: | 1999-05-26 | | Release date: | 1999-11-17 | | Last modified: | 2021-11-03 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Roles of catalytic residues in alpha-amylases as evidenced by the structures of the product-complexed mutants of a maltotetraose-forming amylase.
Protein Eng., 12, 1999
|
|
1QI4
 
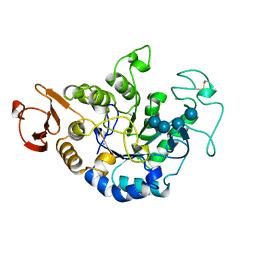 | | MUTANT (E219G) MALTOTETRAOSE-FORMING EXO-AMYLASE IN COMPLEX WITH MALTOTETRAOSE | | Descriptor: | CALCIUM ION, PROTEIN (EXO-MALTOTETRAOHYDROLASE), alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose | | Authors: | Hasegawa, K, Kubota, M, Matsuura, Y. | | Deposit date: | 1999-06-01 | | Release date: | 1999-11-24 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Roles of catalytic residues in alpha-amylases as evidenced by the structures of the product-complexed mutants of a maltotetraose-forming amylase.
Protein Eng., 12, 1999
|
|
