1LBA
 
 | |
1PVI
 
 | | STRUCTURE OF PVUII ENDONUCLEASE WITH COGNATE DNA | | Descriptor: | DNA (5'-D(*TP*GP*AP*CP*CP*AP*GP*CP*TP*GP*GP*TP*C)-3'), PROTEIN (PVUII (E.C.3.1.21.4)) | | Authors: | Cheng, X, Balendiran, K, Schildkraut, I, Anderson, J.E. | | Deposit date: | 1994-11-16 | | Release date: | 1995-02-14 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structure of PvuII endonuclease with cognate DNA.
EMBO J., 13, 1994
|
|
5CX3
 
 | | Crystal structure of FYCO1 LIR in complex with LC3A | | Descriptor: | FYVE and coiled-coil domain-containing protein 1, GLYCEROL, Microtubule-associated proteins 1A/1B light chain 3A | | Authors: | Cheng, X, Pan, L. | | Deposit date: | 2015-07-28 | | Release date: | 2016-08-03 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structural basis of FYCO1 and MAP1LC3A interaction reveals a novel binding mode for Atg8-family proteins.
Autophagy, 12, 2016
|
|
8YRO
 
 | | SARS-CoV-2 Delta Spike in complex with JL-8C | | Descriptor: | JL-8C Heavy Chain, JL-8C Light Chain, Spike glycoprotein | | Authors: | Nguyen, V.H.T, Chen, X. | | Deposit date: | 2024-03-21 | | Release date: | 2024-06-05 | | Last modified: | 2024-06-26 | | Method: | ELECTRON MICROSCOPY (3.27 Å) | | Cite: | The presence of broadly neutralizing anti-SARS-CoV-2 RBD antibodies elicited by primary series and booster dose of COVID-19 vaccine.
Plos Pathog., 20, 2024
|
|
4TYB
 
 | | An Ligand-observed Mass Spectrometry-based Approach Integrated into the Fragment Based Lead Discovery Pipeline | | Descriptor: | (2R)-morpholin-4-yl(phenyl)ethanenitrile, Polyprotein | | Authors: | Shui, W, Yang, C, Lin, J, Chen, X, Qin, S, Chen, S. | | Deposit date: | 2014-07-08 | | Release date: | 2015-05-20 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.93 Å) | | Cite: | A ligand-observed mass spectrometry approach integrated into the fragment based lead discovery pipeline
Sci Rep, 5, 2015
|
|
4TY9
 
 | | An Ligand-observed Mass Spectrometry-based Approach Integrated into the Fragment Based Lead Discovery Pipeline | | Descriptor: | 5-(trifluoromethyl)pyridin-2-amine, Polyprotein | | Authors: | Shui, W, Yang, C, Lin, J, Chen, X, Qin, S, Chen, S. | | Deposit date: | 2014-07-08 | | Release date: | 2015-05-06 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.78 Å) | | Cite: | A ligand-observed mass spectrometry approach integrated into the fragment based lead discovery pipeline
Sci Rep, 5, 2015
|
|
4TYA
 
 | | An Ligand-observed Mass Spectrometry-based Approach Integrated into the Fragment Based Lead Discovery Pipeline | | Descriptor: | 4-(trifluoromethyl)benzoic acid, Polyprotein | | Authors: | Shui, W, Yang, C, Lin, J, Chen, X, Qin, S, Chen, S. | | Deposit date: | 2014-07-08 | | Release date: | 2015-05-06 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.94 Å) | | Cite: | A ligand-observed mass spectrometry approach integrated into the fragment based lead discovery pipeline
Sci Rep, 5, 2015
|
|
4TXS
 
 | | An Ligand-observed Mass Spectrometry-based Approach Integrated into the Fragment Based Lead Discovery Pipeline | | Descriptor: | (4-hydroxyphenyl)acetonitrile, Polyprotein | | Authors: | Shui, W, Yang, C, Lin, J, Chen, X, Qin, S, Chen, S. | | Deposit date: | 2014-07-07 | | Release date: | 2015-05-06 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.78 Å) | | Cite: | A ligand-observed mass spectrometry approach integrated into the fragment based lead discovery pipeline
Sci Rep, 5, 2015
|
|
4TY8
 
 | | An Ligand-observed Mass Spectrometry-based Approach Integrated into the Fragment Based Lead Discovery Pipeline | | Descriptor: | 6-methyl-2H-chromen-2-one, Polyprotein | | Authors: | Shui, W, Yang, C, Lin, J, Chen, X, Qin, S, Chen, S. | | Deposit date: | 2014-07-08 | | Release date: | 2015-05-06 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.78 Å) | | Cite: | A ligand-observed mass spectrometry approach integrated into the fragment based lead discovery pipeline
Sci Rep, 5, 2015
|
|
6DUP
 
 | | CRYSTAL STRUCTURE OF PXR IN COMPLEX WITH COMPOUND 7 | | Descriptor: | (2S)-2-({[3'-(trifluoromethyl)[1,1'-biphenyl]-4-yl]oxy}methyl)-2,3-dihydro-7H-[1,3]oxazolo[3,2-a]pyrimidin-7-one, Nuclear receptor subfamily 1 group I member 2 | | Authors: | Chen, X, Zhang, Y, Mclean, L.R. | | Deposit date: | 2018-06-21 | | Release date: | 2018-08-29 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Amelioration of PXR-mediated CYP3A4 induction by mGluR2 modulators.
Bioorg. Med. Chem. Lett., 28, 2018
|
|
7BTK
 
 | | E.coli beta-galactosidase (E537Q) in complex with fluorescent probe KSA01 | | Descriptor: | 4-[[2-[(E)-2-[4-[(2S,3R,4S,5R,6R)-6-(hydroxymethyl)-3,4,5-tris(oxidanyl)oxan-2-yl]oxyphenyl]ethenyl]-3,3-dimethyl-2H-indol-1-yl]methyl]benzoic acid, Beta-galactosidase, DIMETHYL SULFOXIDE, ... | | Authors: | Chen, X, Hu, Y.L, Liu, Q.M, Gao, Y, Yuan, R, Guo, Y. | | Deposit date: | 2020-04-01 | | Release date: | 2021-03-03 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Two-Dimensional Design Strategy to Construct Smart Fluorescent Probes for the Precise Tracking of Senescence.
Angew.Chem.Int.Ed.Engl., 60, 2021
|
|
7BRS
 
 | | E.coli beta-galactosidase (E537Q) in complex with fluorescent probe KSA02 | | Descriptor: | 8-[2-[(E)-2-[4-[(2S,3R,4S,5R,6R)-6-(hydroxymethyl)-3,4,5-tris(oxidanyl)oxan-2-yl]oxyphenyl]ethenyl]-3,3-dimethyl-indol-1-ium-1-yl]octanoic acid, Beta-galactosidase, DIMETHYL SULFOXIDE, ... | | Authors: | Chen, X, Hu, Y.L, Gao, Y, Yuan, R, Guo, Y. | | Deposit date: | 2020-03-30 | | Release date: | 2021-03-03 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.67 Å) | | Cite: | Two-Dimensional Design Strategy to Construct Smart Fluorescent Probes for the Precise Tracking of Senescence.
Angew.Chem.Int.Ed.Engl., 60, 2021
|
|
6KUZ
 
 | | E.coli beta-galactosidase (E537Q) in complex with fluorescent probe KSL01 | | Descriptor: | 3-(1,3-benzothiazol-2-yl)-2-[[4-[(2~{S},3~{R},4~{S},5~{R},6~{R})-6-(hydroxymethyl)-3,4,5-tris(oxidanyl)oxan-2-yl]oxyphenyl]methoxy]-5-methyl-benzaldehyde, Beta-galactosidase, DIMETHYL SULFOXIDE, ... | | Authors: | Chen, X, Hu, Y.L, Li, X.K, Guo, Y, Li, J. | | Deposit date: | 2019-09-03 | | Release date: | 2020-07-08 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.83 Å) | | Cite: | First-generation species-selective chemical probes for fluorescence imaging of human senescence-associated beta-galactosidase.
Chem Sci, 11, 2020
|
|
7XYO
 
 | | Crystal Structure of a M61 aminopeptidase family member from Myxococcus fulvus | | Descriptor: | Aminopeptidase M61, HEXAETHYLENE GLYCOL, TETRAETHYLENE GLYCOL, ... | | Authors: | Chen, X, Wang, X, Huo, L, Wu, D. | | Deposit date: | 2022-06-02 | | Release date: | 2023-06-07 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Discovery and Characterization of a Myxobacterial Lanthipeptide with Unique Biosynthetic Features and Anti-inflammatory Activity.
J.Am.Chem.Soc., 145, 2023
|
|
1WT8
 
 | | Solution Structure of BmP08 from the Venom of Scorpion Buthus martensii Karsch, 20 structures | | Descriptor: | Neurotoxin BmK X | | Authors: | Wu, H, Chen, X, Tong, X, Li, Y, Zhang, N, Wu, G. | | Deposit date: | 2004-11-17 | | Release date: | 2005-04-19 | | Last modified: | 2022-03-02 | | Method: | SOLUTION NMR | | Cite: | Solution structure of BmP08, a novel short-chain scorpion toxin from Buthus martensi Karsch.
Biochem.Biophys.Res.Commun., 330, 2005
|
|
1BF5
 
 | | TYROSINE PHOSPHORYLATED STAT-1/DNA COMPLEX | | Descriptor: | DNA (5'-D(*AP*CP*AP*GP*TP*TP*TP*CP*CP*CP*GP*TP*AP*AP*AP*TP*G P*C)-3'), DNA (5'-D(*TP*GP*CP*AP*TP*TP*TP*AP*CP*GP*GP*GP*AP*AP*AP*CP*T P*G)-3'), SIGNAL TRANSDUCER AND ACTIVATOR OF TRANSCRIPTION 1-ALPHA/BETA | | Authors: | Kuriyan, J, Zhao, Y, Chen, X, Vinkemeier, U, Jeruzalmi, D, Darnell Jr, J.E. | | Deposit date: | 1998-05-27 | | Release date: | 1998-08-12 | | Last modified: | 2022-12-21 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Crystal structure of a tyrosine phosphorylated STAT-1 dimer bound to DNA.
Cell(Cambridge,Mass.), 93, 1998
|
|
3VFE
 
 | | Virtual Screening and X-Ray Crystallography for Human Kallikrein 6 Inhibitors with an Amidinothiophene P1 Group | | Descriptor: | 4-{[(3R)-3-{[(7-methoxynaphthalen-2-yl)sulfonyl](thiophen-3-ylmethyl)amino}-2-oxopyrrolidin-1-yl]methyl}thiophene-2-carboximidamide, Kallikrein-6 | | Authors: | Chen, X, Zhang, Y, Xia, T, Wang, R. | | Deposit date: | 2012-01-09 | | Release date: | 2012-11-21 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (1.88 Å) | | Cite: | Virtual Screening and X-ray Crystallography for Human Kallikrein 6 Inhibitors with an Amidinothiophene P1 Group.
Acs Med.Chem.Lett., 3, 2012
|
|
2Z59
 
 | | Complex Structures of Mouse Rpn13 (22-130aa) and ubiquitin | | Descriptor: | Protein ADRM1, Ubiquitin | | Authors: | Chen, X, Schreiner, P, Groll, M, Walters, K.J. | | Deposit date: | 2007-07-01 | | Release date: | 2008-05-20 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Ubiquitin docking at the proteasome through a novel pleckstrin-homology domain interaction.
Nature, 453, 2008
|
|
358D
 
 | |
4YIR
 
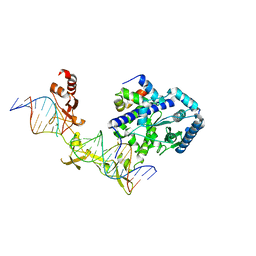 | | Crystal structure of Rad4-Rad23 crosslinked to an undamaged DNA | | Descriptor: | DNA (5'-D(*AP*TP*TP*GP*TP*AP*GP*CP*G*GP*GP*GP*AP*TP*GP*TP*CP*GP*AP*GP*TP*CP*A)-3'), DNA (5'-D(*TP*TP*GP*AP*CP*TP*CP*(G47)P*AP*CP*AP*TP*CP*CP*CP*CP*CP*GP*CP*TP*AP*CP*AP*A)-3'), DNA repair protein RAD4, ... | | Authors: | Min, J.-H, Chen, X, Kim, Y. | | Deposit date: | 2015-03-02 | | Release date: | 2015-03-11 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (3.0501 Å) | | Cite: | Kinetic gating mechanism of DNA damage recognition by Rad4/XPC.
Nat Commun, 6, 2015
|
|
7XUP
 
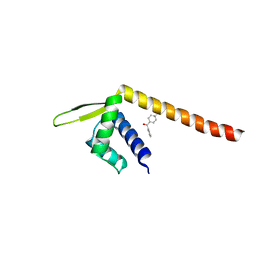 | | Crystal structure of TPe3.0 | | Descriptor: | Transcriptional regulator, PadR-like family | | Authors: | Chen, X, Qian, J.Y, Sun, N.N, Zhong, F.R, Wu, Y.Z. | | Deposit date: | 2022-05-19 | | Release date: | 2022-09-28 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.602 Å) | | Cite: | Enantioselective [2+2]-cycloadditions with triplet photoenzymes.
Nature, 611, 2022
|
|
7XUQ
 
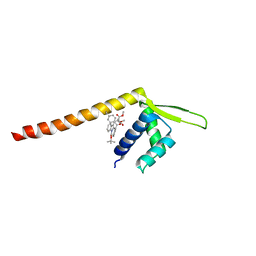 | | Crystal structure of Tpe3.0 complexed with N-Boc-3-alkenylindole | | Descriptor: | Transcriptional regulator, PadR-like family, dimethyl 2-[[2-methyl-1-[(2-methylpropan-2-yl)oxycarbonyl]indol-3-yl]methyl]-2-prop-2-enyl-propanedioate | | Authors: | Chen, X, Qian, J.Y, Sun, N.N, Zhong, F.R, Wu, Y.Z. | | Deposit date: | 2022-05-19 | | Release date: | 2022-09-28 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Enantioselective [2+2]-cycloadditions with triplet photoenzymes.
Nature, 611, 2022
|
|
8X0X
 
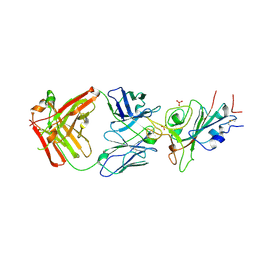 | | Crystal structure of JE-5C in complex with SARS-CoV-2 RBD | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Heavy chain of JE-5C Fab, Light chain of JE-5C Fab, ... | | Authors: | Mohapatra, A, Chen, X. | | Deposit date: | 2023-11-06 | | Release date: | 2024-06-05 | | Last modified: | 2024-07-03 | | Method: | X-RAY DIFFRACTION (3.45 Å) | | Cite: | The presence of broadly neutralizing anti-SARS-CoV-2 RBD antibodies elicited by primary series and booster dose of COVID-19 vaccine.
Plos Pathog., 20, 2024
|
|
8X0Y
 
 | | Crystal structure of JM-1A in complex with SARS-CoV-2 RBD | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, CHLORIDE ION, Heavy chain of JM-1A Fab, ... | | Authors: | Mohapatra, A, Chen, X. | | Deposit date: | 2023-11-06 | | Release date: | 2024-06-05 | | Last modified: | 2024-07-03 | | Method: | X-RAY DIFFRACTION (1.94 Å) | | Cite: | The presence of broadly neutralizing anti-SARS-CoV-2 RBD antibodies elicited by primary series and booster dose of COVID-19 vaccine.
Plos Pathog., 20, 2024
|
|
8YRP
 
 | | SARS-CoV-2 Delta Spike in complex with JM-1A | | Descriptor: | JM-1A Heavy Chain, JM-1A Light Chain, Spike glycoprotein | | Authors: | Nguyen, V.H.T, Chen, X. | | Deposit date: | 2024-03-21 | | Release date: | 2024-06-05 | | Last modified: | 2024-06-26 | | Method: | ELECTRON MICROSCOPY (3.64 Å) | | Cite: | The presence of broadly neutralizing anti-SARS-CoV-2 RBD antibodies elicited by primary series and booster dose of COVID-19 vaccine.
Plos Pathog., 20, 2024
|
|
