5LSY
 
 | | Structure of the Epigenetic Oncogene MMSET and inhibition by N-Alkyl Sinefungin Derivatives | | Descriptor: | Histone-lysine N-methyltransferase SETD2, THIOCYANATE ION, ZINC ION, ... | | Authors: | Tisi, D, Pathuri, P, Heightman, T. | | Deposit date: | 2016-09-05 | | Release date: | 2016-10-12 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.62 Å) | | Cite: | Structure of the Epigenetic Oncogene MMSET and Inhibition by N-Alkyl Sinefungin Derivatives.
ACS Chem. Biol., 11, 2016
|
|
6RRW
 
 | | GOLGI ALPHA-MANNOSIDASE II in complex with (2R,3R,4R,5S)-1-(5-{[4-(3,4-Dihydro-2H-1,5-benzodioxepin-7-yl)benzyl]oxy}pentyl)-2-(hydroxymethyl)-3,4,5-piperidinetriol | | Descriptor: | (2~{R},3~{R},4~{R},5~{S})-1-[5-[[4-(3,4-dihydro-2~{H}-1,5-benzodioxepin-7-yl)phenyl]methoxy]pentyl]-2-(hydroxymethyl)piperidine-3,4,5-triol, Alpha-mannosidase 2, ZINC ION | | Authors: | Armstrong, Z, Lahav, D, Johnson, R, Kuo, C.L, Beenakker, T.J.M, de Boer, C, Wong, C.S, van Rijssel, E.R, Debets, M, Geurink, P.P, Ovaa, H, van der Stelt, M, Codee, J.D.C, Aerts, J.M.F.G, Wu, L, Overkleeft, H.S, Davies, G.J. | | Deposit date: | 2019-05-20 | | Release date: | 2020-07-08 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Manno- epi -cyclophellitols Enable Activity-Based Protein Profiling of Human alpha-Mannosidases and Discovery of New Golgi Mannosidase II Inhibitors.
J.Am.Chem.Soc., 142, 2020
|
|
1LXL
 
 | | NMR STRUCTURE OF BCL-XL, AN INHIBITOR OF PROGRAMMED CELL DEATH, MINIMIZED AVERAGE STRUCTURE | | Descriptor: | BCL-XL | | Authors: | Muchmore, S.W, Sattler, M, Liang, H, Meadows, R.P, Harlan, J.E, Yoon, H.S, Nettesheim, D, Chang, B.S, Thompson, C.B, Wong, S.L, Ng, S.C, Fesik, S.W. | | Deposit date: | 1996-04-04 | | Release date: | 1997-04-21 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | X-ray and NMR structure of human Bcl-xL, an inhibitor of programmed cell death.
Nature, 381, 1996
|
|
5LT6
 
 | | Structure of the Epigenetic Oncogene MMSET and inhibition by N-Alkyl Sinefungin Derivatives | | Descriptor: | Histone-lysine N-methyltransferase SETD2, THIOCYANATE ION, ZINC ION, ... | | Authors: | Tisi, D, Pathuri, P, Heightman, T. | | Deposit date: | 2016-09-06 | | Release date: | 2016-10-05 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Structure of the Epigenetic Oncogene MMSET and Inhibition by N-Alkyl Sinefungin Derivatives.
ACS Chem. Biol., 11, 2016
|
|
3VVW
 
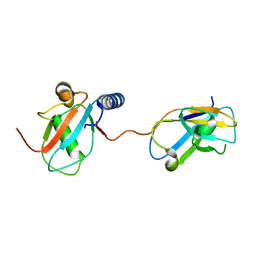 | | NDP52 in complex with LC3C | | Descriptor: | Calcium-binding and coiled-coil domain-containing protein 2, Microtubule-associated proteins 1A/1B light chain 3C | | Authors: | Akutsu, M, Muhlinen, N.V, Randow, F, Komander, D. | | Deposit date: | 2012-07-28 | | Release date: | 2013-02-27 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | LC3C, bound selectively by a noncanonical LIR motif in NDP52, is required for antibacterial autophagy
Mol.Cell, 48, 2012
|
|
5LTM
 
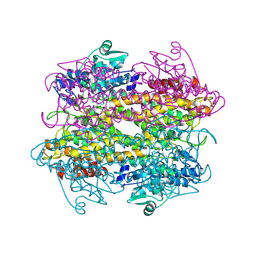 | |
2GFT
 
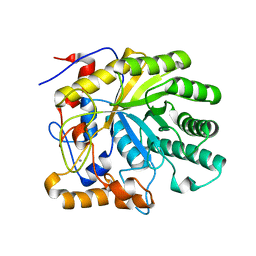 | | Crystal structure of the E263A nucleophile mutant of Bacillus licheniformis endo-beta-1,4-galactanase in complex with galactotriose | | Descriptor: | CALCIUM ION, Glycosyl Hydrolase Family 53, beta-D-galactopyranose-(1-4)-beta-D-galactopyranose-(1-4)-beta-D-galactopyranose | | Authors: | Welner, D, Le Nours, J, De Maria, L, Jorgensen, C.T, Christensen, L.L.H, Larsen, S, Lo Leggio, L. | | Deposit date: | 2006-03-23 | | Release date: | 2006-06-13 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Oligosaccharide binding of Bacillus licheniformis endo-beta-1,4-galactanase
To be Published
|
|
1MFR
 
 | | CRYSTAL STRUCTURE OF M FERRITIN | | Descriptor: | M FERRITIN, MAGNESIUM ION | | Authors: | Ha, Y, Shi, D, Allewell, N.M. | | Deposit date: | 1998-06-18 | | Release date: | 1999-06-22 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Crystal structure of bullfrog M ferritin at 2.8 A resolution: analysis of subunit interactions and the binuclear metal center
J.Biol.Inorg.Chem., 4, 1999
|
|
6S46
 
 | | Room temperature structure of the LOV2 domain of phototropin-2 from Arabidopsis thaliana 4158 ms after initiation of illumination, determined with a serial crystallography approach | | Descriptor: | FLAVIN MONONUCLEOTIDE, Phototropin-2 | | Authors: | Aumonier, S, Santoni, G, Gotthard, G, von Stetten, D, Leonard, G, Royant, A. | | Deposit date: | 2019-06-26 | | Release date: | 2020-07-08 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.75 Å) | | Cite: | Millisecond time-resolved serial oscillation crystallography of a blue-light photoreceptor at a synchrotron.
Iucrj, 7, 2020
|
|
5O61
 
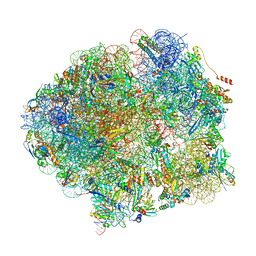 | | The complete structure of the Mycobacterium smegmatis 70S ribosome | | Descriptor: | 16S rRNA, 23S rRNA, 30S ribosomal protein S10, ... | | Authors: | Hentschel, J, Burnside, C, Mignot, I, Leibundgut, M, Boehringer, D, Ban, N. | | Deposit date: | 2017-06-03 | | Release date: | 2017-07-12 | | Last modified: | 2024-05-15 | | Method: | ELECTRON MICROSCOPY (3.31 Å) | | Cite: | The Complete Structure of the Mycobacterium smegmatis 70S Ribosome.
Cell Rep, 20, 2017
|
|
5OC6
 
 | |
1NIX
 
 | |
1NJ8
 
 | | Crystal Structure of Prolyl-tRNA Synthetase from Methanocaldococcus janaschii | | Descriptor: | Proline-tRNA Synthetase | | Authors: | Kamtekar, S, Kennedy, W.D, Wang, J, Stathopoulos, C, Soll, D, Steitz, T.A. | | Deposit date: | 2002-12-30 | | Release date: | 2003-03-04 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | The structural basis of cysteine aminoacylation of tRNAPro by prolyl-tRNA synthetases
Proc.Natl.Acad.Sci.USA, 100, 2003
|
|
2G6V
 
 | | The crystal structure of ribD from Escherichia coli | | Descriptor: | Riboflavin biosynthesis protein ribD | | Authors: | Stenmark, P, Moche, M, Gurmu, D, Nordlund, P, Structural Proteomics in Europe (SPINE) | | Deposit date: | 2006-02-25 | | Release date: | 2007-02-06 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | The Crystal Structure of the Bifunctional Deaminase/Reductase RibD of the Riboflavin Biosynthetic Pathway in Escherichia coli: Implications for the Reductive Mechanism.
J.Mol.Biol., 373, 2007
|
|
6RKA
 
 | | Inter-dimeric interface controls function and stability of S-methionine adenosyltransferase from U. urealiticum | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, Methionine adenosyltransferase, PHOSPHATE ION, ... | | Authors: | Shahar, A, Zarivach, R, Bershtein, S, Kleiner, D, Shmulevich, F. | | Deposit date: | 2019-04-30 | | Release date: | 2019-09-25 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | The interdimeric interface controls function and stability of Ureaplasma urealiticum methionine S-adenosyltransferase.
J.Mol.Biol., 431, 2019
|
|
2GF0
 
 | | The crystal structure of the human DiRas1 GTPase in the inactive GDP bound state | | Descriptor: | GTP-binding protein Di-Ras1, GUANOSINE-5'-DIPHOSPHATE, MAGNESIUM ION | | Authors: | Turnbull, A.P, Papagrigoriou, E, Yang, X, Schoch, G, Elkins, J, Gileadi, O, Salah, E, Bray, J, Wen-Hwa, L, Fedorov, O, Niesen, F.E, von Delft, F, Weigelt, J, Edwards, A, Arrowsmith, C, Sundstrom, M, Doyle, D, Structural Genomics Consortium (SGC) | | Deposit date: | 2006-03-21 | | Release date: | 2006-04-04 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | The crystal structure of the human DiRas1 GTPase in the inactive GDP bound state
To be Published
|
|
6T10
 
 | | Crystal structure of YTHDC1 with fragment 28 (DHU_DC1_176) | | Descriptor: | 5-iodanyl-~{N}-methyl-1~{H}-indazole-3-carboxamide, SULFATE ION, YTHDC1 | | Authors: | Bedi, R.K, Huang, D, Sledz, P, Caflisch, A. | | Deposit date: | 2019-10-03 | | Release date: | 2020-03-04 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.48 Å) | | Cite: | Selectively Disrupting m6A-Dependent Protein-RNA Interactions with Fragments.
Acs Chem.Biol., 15, 2020
|
|
2GA6
 
 | | The crystal structure of SARS nsp10 without zinc ion as additive | | Descriptor: | ZINC ION, orf1a polyprotein | | Authors: | Su, D, Lou, Z, Sun, F, Zhai, Y, Yang, H, Rao, Z. | | Deposit date: | 2006-03-08 | | Release date: | 2006-08-15 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Dodecamer Structure of Severe Acute Respiratory Syndrome Coronavirus Nonstructural Protein nsp10
J.Virol., 80, 2006
|
|
2GI4
 
 | |
5NUP
 
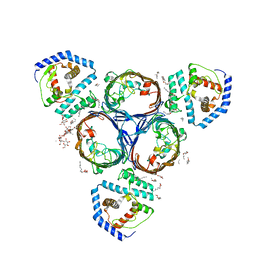 | | Structural basis for maintenance of bacterial outer membrane lipid asymmetry | | Descriptor: | (HYDROXYETHYLOXY)TRI(ETHYLOXY)OCTANE, ABC transporter permease, DODECYL-BETA-D-MALTOSIDE, ... | | Authors: | Abellon-Ruiz, J, Kaptan, S.S, Basle, A, Claudi, B, Bumann, D, Kleinekathofer, U, van den Berg, B. | | Deposit date: | 2017-05-01 | | Release date: | 2017-10-25 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Structural basis for maintenance of bacterial outer membrane lipid asymmetry.
Nat Microbiol, 2, 2017
|
|
6SK6
 
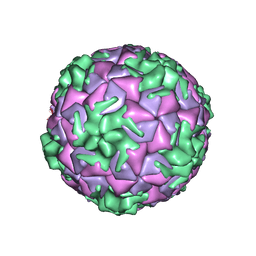 | | Cryo-EM structure of rhinovirus-B5 | | Descriptor: | Rhinovirus B5 VP1, Rhinovirus B5 VP2, Rhinovirus B5 VP3, ... | | Authors: | Wald, J, Goessweiner-Mohr, N, Blaas, D, Pasin, M. | | Deposit date: | 2019-08-14 | | Release date: | 2019-09-04 | | Last modified: | 2024-05-22 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Cryo-EM structure of pleconaril-resistant rhinovirus-B5 complexed to the antiviral OBR-5-340 reveals unexpected binding site.
Proc.Natl.Acad.Sci.USA, 116, 2019
|
|
5NUQ
 
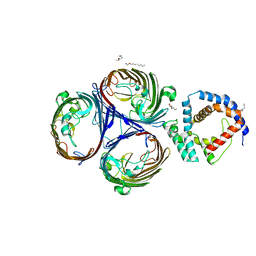 | | Structural basis for maintenance of bacterial outer membrane lipid asymmetry | | Descriptor: | (HYDROXYETHYLOXY)TRI(ETHYLOXY)OCTANE, Outer membrane protein F, Probable phospholipid-binding lipoprotein mlaA | | Authors: | Abellon-Ruiz, J, Kaptan, S.S, Basle, A, Claudi, B, Bumann, D, Kleinekathofer, U, van den Berg, B. | | Deposit date: | 2017-05-01 | | Release date: | 2017-10-25 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Structural basis for maintenance of bacterial outer membrane lipid asymmetry.
Nat Microbiol, 2, 2017
|
|
1NI0
 
 | |
6SKO
 
 | | Cryo-EM Structure of the Fork Protection Complex Bound to CMG at a Replication Fork - conformation 2 MCM CTD:ssDNA | | Descriptor: | DNA replication licensing factor MCM2, DNA replication licensing factor MCM3, DNA replication licensing factor MCM4, ... | | Authors: | Yeeles, J, Baretic, D, Jenkyn-Bedford, M. | | Deposit date: | 2019-08-16 | | Release date: | 2020-05-06 | | Last modified: | 2024-05-22 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Cryo-EM Structure of the Fork Protection Complex Bound to CMG at a Replication Fork.
Mol.Cell, 78, 2020
|
|
1NIP
 
 | | CRYSTALLOGRAPHIC STRUCTURE OF THE NITROGENASE IRON PROTEIN FROM AZOTOBACTER VINELANDII | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, IRON/SULFUR CLUSTER, MAGNESIUM ION, ... | | Authors: | Komiya, H, Georgiadis, M.M, Chakrabarti, P, Woo, D, Kornuc, J.J, Rees, D.C. | | Deposit date: | 1992-09-29 | | Release date: | 1993-10-31 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Crystallographic structure of the nitrogenase iron protein from Azotobacter vinelandii.
Science, 257, 1992
|
|
