1PAV
 
 | | SOLUTION NMR STRUCTURE OF HYPOTHETICAL PROTEIN TA1414 OF THERMOPLASMA ACIDOPHILUM | | Descriptor: | Hypothetical protein Ta1170/Ta1414 | | Authors: | Monleon, D, Yee, A, Liu, C.S, Arrowsmith, C, Celda, B. | | Deposit date: | 2003-05-14 | | Release date: | 2003-05-27 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Solution structure of hypothetical protein TA1414 from Thermoplasma acidophilum.
J.Biomol.Nmr, 28, 2004
|
|
6S20
 
 | | Metabolism of multiple glycosaminoglycans by bacteroides thetaiotaomicron is orchestrated by a versatile core genetic locus (BT33336S-sulf) | | Descriptor: | 2-acetamido-2-deoxy-6-O-sulfo-beta-D-galactopyranose, CALCIUM ION, N-acetylgalactosamine-6-O-sulfatase, ... | | Authors: | Ndeh, D, Basle, A, Strahl, H, Henrissat, B, Terrapon, N, Cartmell, A. | | Deposit date: | 2019-06-19 | | Release date: | 2020-02-05 | | Last modified: | 2024-05-15 | | Method: | X-RAY DIFFRACTION (1.98 Å) | | Cite: | Metabolism of multiple glycosaminoglycans by Bacteroides thetaiotaomicron is orchestrated by a versatile core genetic locus.
Nat Commun, 11, 2020
|
|
8F0V
 
 | | Lipocalin-like Milk protein-2 - E38A mutant | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Milk protein, ZINC ION | | Authors: | Subramanian, R, KanagaVijayan, D. | | Deposit date: | 2022-11-04 | | Release date: | 2023-08-23 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.951 Å) | | Cite: | Variability in phenylalanine side chain conformations facilitates broad substrate tolerance of fatty acid binding in cockroach milk proteins.
Plos One, 18, 2023
|
|
8F0Y
 
 | | Lipocalin-like Milk protein-1 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Milk protein | | Authors: | Subramanian, R, KanagaVijayan, D, Shantakumar, R.P.S. | | Deposit date: | 2022-11-04 | | Release date: | 2023-08-23 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Variability in phenylalanine side chain conformations facilitates broad substrate tolerance of fatty acid binding in cockroach milk proteins.
Plos One, 18, 2023
|
|
3UBD
 
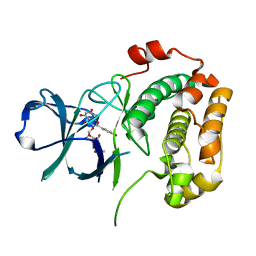 | | Structure of N-terminal domain of RSK2 kinase in complex with flavonoid glycoside SL0101 | | Descriptor: | 5,7-dihydroxy-2-(4-hydroxyphenyl)-4-oxo-4H-chromen-3-yl 3,4-di-O-acetyl-6-deoxy-alpha-L-mannopyranoside, Ribosomal protein S6 kinase alpha-3 | | Authors: | Utepbergenov, D, Derewenda, U, Derewenda, Z.S. | | Deposit date: | 2011-10-24 | | Release date: | 2012-09-05 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.53 Å) | | Cite: | Insights into the Inhibition of the p90 Ribosomal S6 Kinase (RSK) by the Flavonol Glycoside SL0101 from the 1.5 A Crystal Structure of the N-Terminal Domain of RSK2 with Bound Inhibitor.
Biochemistry, 51, 2012
|
|
6RJS
 
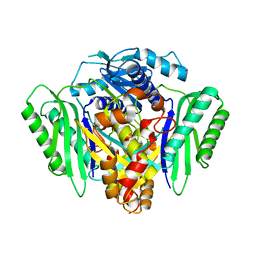 | | Inter-dimeric interface controls function and stability of S-methionine adenosyltransferase from U. urealiticum | | Descriptor: | Methionine adenosyltransferase | | Authors: | Shahar, A, Zarivach, R, Bershtein, S, Kleiner, D, Shmulevich, F. | | Deposit date: | 2019-04-29 | | Release date: | 2019-09-25 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | The interdimeric interface controls function and stability of Ureaplasma urealiticum methionine S-adenosyltransferase.
J.Mol.Biol., 431, 2019
|
|
6RV4
 
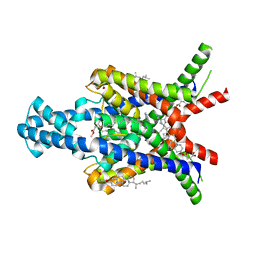 | | Crystal structure of the human two pore domain potassium ion channel TASK-1 (K2P3.1) in a closed conformation with a bound inhibitor BAY 2341237 | | Descriptor: | 1,2-DIACYL-SN-GLYCERO-3-PHOSPHOCHOLINE, CHOLESTEROL HEMISUCCINATE, POTASSIUM ION, ... | | Authors: | Rodstrom, K.E.J, Pike, A.C.W, Zhang, W, Quigley, A, Speedman, D, Mukhopadhyay, S.M.M, Shrestha, L, Chalk, R, Venkaya, S, Bushell, S.R, Tessitore, A, Burgess-Brown, N, Arrowsmith, C.H, Edwards, A.M, Bountra, C, Carpenter, E.P, Structural Genomics Consortium (SGC) | | Deposit date: | 2019-05-30 | | Release date: | 2019-08-07 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | A lower X-gate in TASK channels traps inhibitors within the vestibule.
Nature, 582, 2020
|
|
6RVR
 
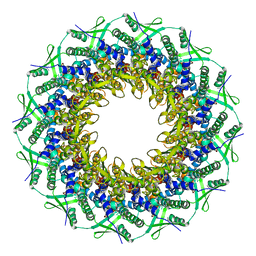 | | Atomic structure of the Epstein-Barr portal, structure I | | Descriptor: | Portal protein | | Authors: | Machon, C, Fabrega-Ferrer, M, Zhou, D, Cuervo, A, Carrascosa, J.L, Stuart, D.I, Coll, M. | | Deposit date: | 2019-05-31 | | Release date: | 2019-09-18 | | Last modified: | 2024-05-22 | | Method: | ELECTRON MICROSCOPY (3.46 Å) | | Cite: | Atomic structure of the Epstein-Barr virus portal.
Nat Commun, 10, 2019
|
|
3U8J
 
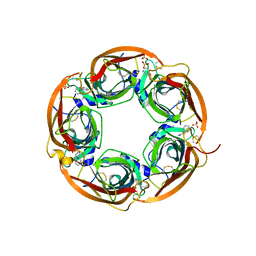 | | Crystal structure of the acetylcholine binding protein (AChBP) from Lymnaea stagnalis in complex with NS3531 (1-(pyridin-3-yl)-1,4-diazepane) | | Descriptor: | 1-(pyridin-3-yl)-1,4-diazepane, 2-acetamido-2-deoxy-beta-D-glucopyranose, Acetylcholine-binding protein, ... | | Authors: | Rohde, L.A.H, Ahring, P.K, Jensen, M.L, Nielsen, E.O, Peters, D, Helgstrand, C, Krintel, C, Harpsoe, K, Gajhede, M, Kastrup, J.S, Balle, T. | | Deposit date: | 2011-10-17 | | Release date: | 2011-12-14 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Intersubunit bridge formation governs agonist efficacy at nicotinic acetylcholine alpha 4 beta 2 receptors: unique role of halogen bonding revealed.
J.Biol.Chem., 287, 2012
|
|
1PES
 
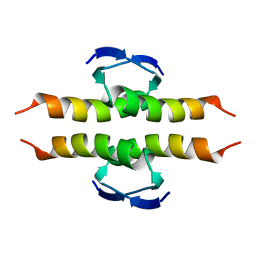 | | NMR SOLUTION STRUCTURE OF THE TETRAMERIC MINIMUM TRANSFORMING DOMAIN OF P53 | | Descriptor: | TUMOR SUPPRESSOR P53 | | Authors: | Lee, W, Harvey, T.S, Yin, Y, Yau, P, Litchfield, D, Arrowsmith, C.H. | | Deposit date: | 1994-11-24 | | Release date: | 1995-02-07 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the tetrameric minimum transforming domain of p53.
Nat.Struct.Biol., 1, 1994
|
|
3U9D
 
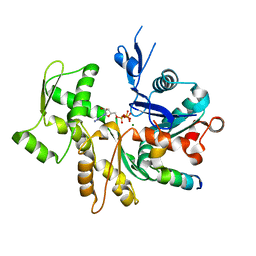 | | Crystal Structure of a chimera containing the N-terminal domain (residues 8-24) of drosophila Ciboulot and the C-terminal domain (residues 13-44) of bovine Thymosin-beta4, bound to G-actin-ATP | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, Actin, alpha skeletal muscle, ... | | Authors: | Renault, L, Husson, C, Carlier, M.F, Didry, D. | | Deposit date: | 2011-10-18 | | Release date: | 2012-01-25 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | How a single residue in individual beta-thymosin/WH2 domains controls their functions in actin assembly.
Embo J., 31, 2012
|
|
5NB4
 
 | | Atomic resolution structure of C-phycoerythrin from marine cyanobacterium Phormidium sp. A09DM at pH 7.5 | | Descriptor: | (4R)-2-METHYLPENTANE-2,4-DIOL, (4S)-2-METHYL-2,4-PENTANEDIOL, HYDROGENPHOSPHATE ION, ... | | Authors: | Sonani, R.R, Roszak, A.W, Ortmann de Percin Northumberland, C, Madamwar, D, Cogdell, R.J. | | Deposit date: | 2017-03-01 | | Release date: | 2017-09-27 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.14 Å) | | Cite: | An improved crystal structure of C-phycoerythrin from the marine cyanobacterium Phormidium sp. A09DM.
Photosyn. Res., 135, 2018
|
|
1PBH
 
 | |
1PB5
 
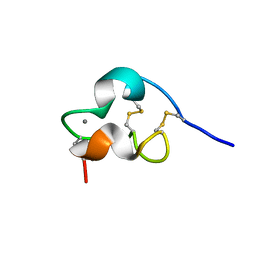 | | NMR Structure of a Prototype LNR Module from Human Notch1 | | Descriptor: | CALCIUM ION, Neurogenic locus notch homolog protein 1 | | Authors: | Vardar, D, North, C.L, Sanchez-Irizarry, C, Aster, J.C, Blacklow, S.C. | | Deposit date: | 2003-05-14 | | Release date: | 2003-06-17 | | Last modified: | 2024-10-09 | | Method: | SOLUTION NMR | | Cite: | Nuclear Magnetic Resonance Structure of a Prototype Lin12-Notch Repeat Module from Human Notch1
Biochemistry, 42, 2003
|
|
5O71
 
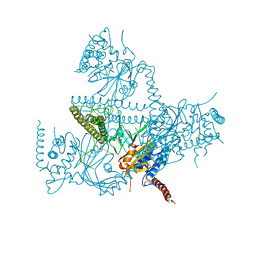 | | Crystal structure of human USP25 | | Descriptor: | Ubiquitin carboxyl-terminal hydrolase 25 | | Authors: | Reverter, D, Liu, B. | | Deposit date: | 2017-06-07 | | Release date: | 2018-06-20 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (3.283 Å) | | Cite: | A quaternary tetramer assembly inhibits the deubiquitinating activity of USP25.
Nat Commun, 9, 2018
|
|
5O7O
 
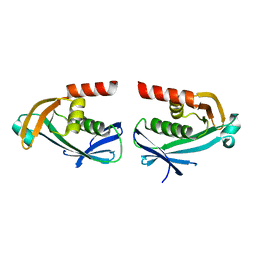 | | The crystal structure of DfoC, the desferrioxamine biosynthetic pathway acetyltransferase/Non-Ribosomal Peptide Synthetase (NRPS)-Independent Siderophore (NIS) from the fire blight disease pathogen Erwinia amylovora | | Descriptor: | Desferrioxamine siderophore biosynthesis protein dfoC | | Authors: | Salomone-Stagni, M, Bartho, J.D, Polsinelli, I, Bellini, D, Walsh, M.A, Demitri, N, Benini, S. | | Deposit date: | 2017-06-09 | | Release date: | 2018-02-28 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.11 Å) | | Cite: | A complete structural characterization of the desferrioxamine E biosynthetic pathway from the fire blight pathogen Erwinia amylovora.
J. Struct. Biol., 202, 2018
|
|
1PG5
 
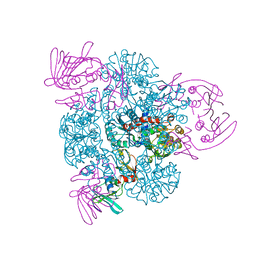 | | CRYSTAL STRUCTURE OF THE UNLIGATED (T-STATE) ASPARTATE TRANSCARBAMOYLASE FROM THE EXTREMELY THERMOPHILIC ARCHAEON SULFOLOBUS ACIDOCALDARIUS | | Descriptor: | Aspartate carbamoyltransferase, Aspartate carbamoyltransferase regulatory chain, ZINC ION | | Authors: | De Vos, D, Van Petegem, F, Remaut, H, Legrain, C, Glansdorff, N, Van Beeumen, J.J. | | Deposit date: | 2003-05-27 | | Release date: | 2004-06-08 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Crystal Structure of T State Aspartate Carbamoyltransferase of the Hyperthermophilic Archaeon Sulfolobus acidocaldarius.
J.Mol.Biol., 339, 2004
|
|
1PJ7
 
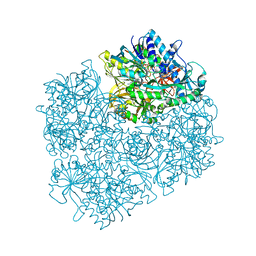 | | Structure of dimethylglycine oxidase of Arthrobacter globiformis in complex with folinic acid | | Descriptor: | FLAVIN-ADENINE DINUCLEOTIDE, N,N-dimethylglycine oxidase, N-[4-({[(6S)-2-amino-5-formyl-4-oxo-3,4,5,6,7,8-hexahydropteridin-6-yl]methyl}amino)benzoyl]-L-glutamic acid, ... | | Authors: | Leys, D, Basran, J, Scrutton, N.S. | | Deposit date: | 2003-06-01 | | Release date: | 2003-10-07 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Channelling and formation of 'active' formaldehyde in dimethylglycine oxidase.
Embo J., 22, 2003
|
|
5O1M
 
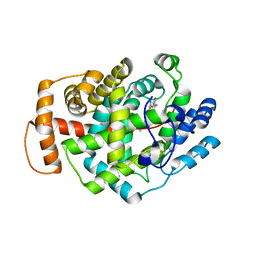 | | Structure of Latex Clearing Protein LCP in the closed state | | Descriptor: | 1,2-ETHANEDIOL, PROTOPORPHYRIN IX CONTAINING FE, Rubber oxygenase | | Authors: | Ilcu, L, Roether, W, Birke, J, Brausemann, A, Einsle, O, Jendrossek, D. | | Deposit date: | 2017-05-18 | | Release date: | 2017-08-02 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structural and Functional Analysis of Latex Clearing Protein (Lcp) Provides Insight into the Enzymatic Cleavage of Rubber.
Sci Rep, 7, 2017
|
|
6TGD
 
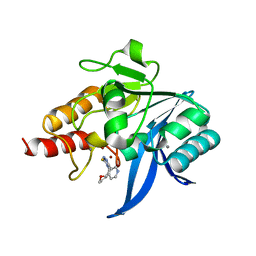 | | Crystal structure of NDM-1 in complex with triazole-based inhibitor OP31 | | Descriptor: | 4-[[(2~{S})-oxolan-2-yl]methyl]-3-pyridin-3-yl-1~{H}-1,2,4-triazole-5-thione, CALCIUM ION, Metallo-beta-lactamase NDM-1, ... | | Authors: | Maso, L, Spirakis, F, Santucci, M, Simon, C, Docquier, J.D, Cruciani, G, Costi, M.P, Tondi, D, Cendron, L. | | Deposit date: | 2019-11-15 | | Release date: | 2020-08-26 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.33 Å) | | Cite: | Virtual screening identifies broad-spectrum beta-lactamase inhibitors with activity on clinically relevant serine- and metallo-carbapenemases.
Sci Rep, 10, 2020
|
|
5O3N
 
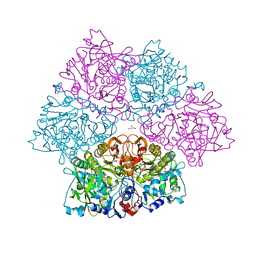 | | Crystal structure of E. cloacae 3,4-dihydroxybenzoic acid decarboxylase (AroY) reconstituted with prFMN | | Descriptor: | 1-deoxy-5-O-phosphono-1-(3,3,4,5-tetramethyl-9,11-dioxo-2,3,8,9,10,11-hexahydro-7H-quinolino[1,8-fg]pteridin-12-ium-7-y l)-D-ribitol, 3,4-dihydroxybenzoate decarboxylase, GLYCEROL, ... | | Authors: | Marshall, S.A, Leys, D. | | Deposit date: | 2017-05-24 | | Release date: | 2017-09-13 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Regioselective para-Carboxylation of Catechols with a Prenylated Flavin Dependent Decarboxylase.
Angew. Chem. Int. Ed. Engl., 56, 2017
|
|
5OD1
 
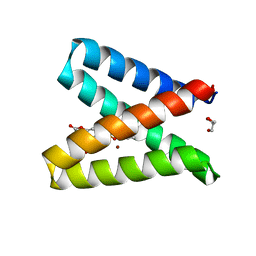 | | Structure of the engineered metalloesterase MID1sc10 complexed with a phosphonate transition state analogue | | Descriptor: | GLYCEROL, MID1sc10, ZINC ION, ... | | Authors: | Mittl, P.R.E, Studer, S, Hansen, D.A, Hilvert, D. | | Deposit date: | 2017-07-04 | | Release date: | 2018-12-12 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.34 Å) | | Cite: | Evolution of a highly active and enantiospecific metalloenzyme from short peptides.
Science, 362, 2018
|
|
5OD8
 
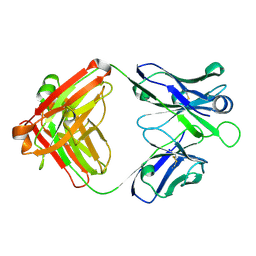 | | Crystal structure of the RA-associated mAb B2 (Fab fragment) | | Descriptor: | B2 Fab fragment - Light chain, B2 Fab fragment - heavy chain | | Authors: | Dobritzsch, D, Ge, C, Holmdahl, R, Amara, K, Malmstrom, V. | | Deposit date: | 2017-07-05 | | Release date: | 2018-07-04 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structural Basis of Cross-Reactivity of Anti-Citrullinated Protein Antibodies.
Arthritis Rheumatol, 71, 2019
|
|
6TE4
 
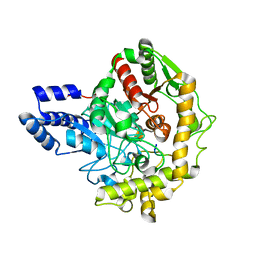 | | Structural insights into Pseudomonas aeruginosa Type six secretion system exported effector 8: Tse8 in complex with a peptide | | Descriptor: | Pro-Pro-Leu-Ala-Ser-Lys, Tse8 | | Authors: | Sainz-Polo, M.A, Capuni, R, Lucas, M, Altuna, J, Fucini, P, Montanchez, I, Albesa-Jove, D. | | Deposit date: | 2019-11-11 | | Release date: | 2020-11-04 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.29 Å) | | Cite: | Structural insights into Pseudomonas aeruginosaType six secretion system exported effector 8.
J.Struct.Biol., 212, 2020
|
|
5O3Z
 
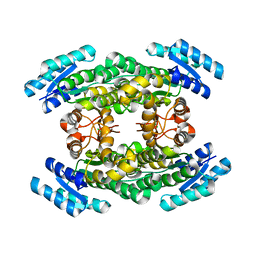 | | Crystal structure of Sorbitol-6-Phosphate 2-dehydrogenase SrlD from Erwinia amylovora | | Descriptor: | CHLORIDE ION, Sorbitol-6-phosphate dehydrogenase | | Authors: | Salomone-Stagni, M, Bartho, J.D, Bellini, D, Walsh, M.A, Benini, S. | | Deposit date: | 2017-05-25 | | Release date: | 2018-06-13 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.84 Å) | | Cite: | Structural and functional analysis of Erwinia amylovora SrlD. The first crystal structure of a sorbitol-6-phosphate 2-dehydrogenase.
J.Struct.Biol., 203, 2018
|
|
