1ZKI
 
 | | Structure of conserved protein PA5202 from Pseudomonas aeruginosa | | Descriptor: | ACETIC ACID, hypothetical protein PA5202 | | Authors: | Cuff, M.E, Evdokimova, E, Edwards, A, Savchenko, A, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2005-05-02 | | Release date: | 2005-06-14 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structure and activity of the Pseudomonas aeruginosa hotdog-fold thioesterases PA5202 and PA2801.
Biochem.J., 444, 2012
|
|
2OKG
 
 | | Structure of effector binding domain of central glycolytic gene regulator (CggR) from B. subtilis | | Descriptor: | CHLORIDE ION, Central glycolytic gene regulator, GLYCERALDEHYDE-3-PHOSPHATE | | Authors: | Rezacova, P, Moy, S.F, Joachimiak, A, Otwinowski, Z, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2007-01-16 | | Release date: | 2007-01-30 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Crystal structures of the effector-binding domain of repressor Central glycolytic gene Regulator from Bacillus subtilis reveal ligand-induced structural changes upon binding of several glycolytic intermediates.
Mol.Microbiol., 69, 2008
|
|
2O3G
 
 | | Structural Genomics, the crystal structure of a conserved putative domain from Neisseria meningitidis MC58 | | Descriptor: | 1,2-ETHANEDIOL, Putative protein | | Authors: | Tan, K, Volkart, L, Gu, M, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2006-12-01 | | Release date: | 2007-01-02 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.55 Å) | | Cite: | The crystal structure of a conserved putative domain from Neisseria meningitidis MC58
To be Published
|
|
1YX1
 
 | | Crystal Structure of Protein of Unknown Function PA2260 from Pseudomonas aeruginosa, Possible Sugar Phosphate Isomerase | | Descriptor: | ISOPROPYL ALCOHOL, SODIUM ION, hypothetical protein PA2260 | | Authors: | Osipiuk, J, Xu, X, Savchenko, A, Edwards, A, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2005-02-19 | | Release date: | 2005-04-05 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | X-ray crystal structure of hypothetical protein PA2260 from Pseudomonas aeruginosa
To be Published
|
|
2ODF
 
 | | The crystal structure of gene product Atu2144 from Agrobacterium tumefaciens | | Descriptor: | Hypothetical protein Atu2144, SULFATE ION | | Authors: | Zhang, R, Xu, X, Zheng, H, Savchenko, A, Edwards, A, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2006-12-22 | | Release date: | 2007-01-23 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | The crystal structure of gene product Atu2144 from Agrobacterium tumefaciens
To be Published
|
|
2O6I
 
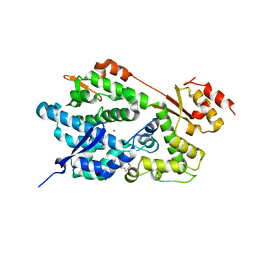 | | Structure of an Enterococcus Faecalis HD Domain Phosphohydrolase | | Descriptor: | 2-HYDROXYETHYL DISULFIDE, CHLORIDE ION, HD domain protein, ... | | Authors: | Vorontsov, I.I, Minasov, G, Shuvalova, L, Brunzelle, J.S, Moy, S, Collart, F.R, Joachimiak, A, Anderson, W.F, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2006-12-07 | | Release date: | 2006-12-19 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.55 Å) | | Cite: | Characterization of the deoxynucleotide triphosphate triphosphohydrolase (dNTPase) activity of the EF1143 protein from Enterococcus faecalis and crystal structure of the activator-substrate complex.
J.Biol.Chem., 286, 2011
|
|
2OQT
 
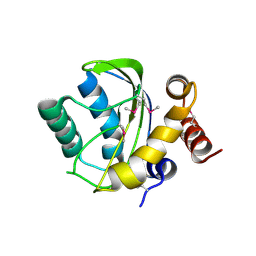 | | Structural Genomics, the crystal structure of a putative PTS IIA domain from Streptococcus pyogenes M1 GAS | | Descriptor: | Hypothetical protein SPy0176 | | Authors: | Tan, K, Wu, R, Osipiuk, J, Clancy, S, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2007-02-01 | | Release date: | 2007-03-06 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.41 Å) | | Cite: | The crystal structure of a putative PTS IIA domain from Streptococcus pyogenes M1 GAS
To be Published
|
|
1YXY
 
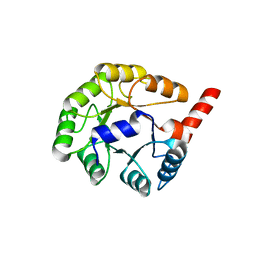 | | Crystal Structure of putative N-acetylmannosamine-6-P epimerase from Streptococcus pyogenes (APC29713) Structural genomics, MCSG | | Descriptor: | Putative N-acetylmannosamine-6-phosphate 2-epimerase | | Authors: | Rotella, F.J, Zhang, R.G, Lezondra, L.E.O, Collart, F.R, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2005-02-22 | | Release date: | 2005-04-05 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | The 1.6 A crystal structure of putative N-acetylmannosamine-6-P epimerase from Streptococcus pyogenes
To be Published
|
|
2AO9
 
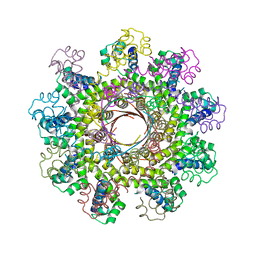 | |
2R6O
 
 | | Crystal structure of putative diguanylate cyclase/phosphodiesterase from Thiobacillus denitrificans | | Descriptor: | CHLORIDE ION, MAGNESIUM ION, Putative diguanylate cyclase/phosphodiesterase (GGDEF & EAL domains) | | Authors: | Chang, C, Xu, X, Zheng, H, Savchenko, A, Edwards, A.M, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2007-09-06 | | Release date: | 2007-09-18 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structural insight into the mechanism of c-di-GMP hydrolysis by EAL domain phosphodiesterases.
J.Mol.Biol., 402, 2010
|
|
2RC3
 
 | | Crystal structure of CBS domain, NE2398 | | Descriptor: | BROMIDE ION, CBS domain, NICOTINAMIDE-ADENINE-DINUCLEOTIDE | | Authors: | Dong, A, Xu, X, Korniyenko, Y, Yakunin, A, Zheng, H, Walker, J.R, Edwards, A.M, Joachimiak, A, Savchenko, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2007-09-19 | | Release date: | 2007-10-02 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Crystal structure of CBS domain, NE2398.
To be Published
|
|
2PQQ
 
 | | Structural Genomics, the crystal structure of the N-terminal domain of a transcriptional regulator from Streptomyces coelicolor A3(2) | | Descriptor: | FORMIC ACID, Putative transcriptional regulator | | Authors: | Tan, K, Xu, X, Zheng, H, Savchenko, A, Edwards, A.M, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2007-05-02 | | Release date: | 2007-06-05 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | The crystal structure of the N-terminal domain of a transcriptional regulator from Streptomyces coelicolor A3(2)
To be Published
|
|
2GYQ
 
 | | YcfI, a putative structural protein from Rhodopseudomonas palustris. | | Descriptor: | 1,2-ETHANEDIOL, ACETATE ION, FE (III) ION, ... | | Authors: | Osipiuk, J, Evdokimova, E, Kudritska, M, Savchenko, A, Edwards, A, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2006-05-09 | | Release date: | 2006-06-13 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | X-ray crystal structure of YcfI protein, a putative structural protein from Rhodopseudomonas palustris.
To be Published
|
|
2QIP
 
 | | Crystal structure of a protein of unknown function VPA0982 from Vibrio parahaemolyticus RIMD 2210633 | | Descriptor: | 1,2-ETHANEDIOL, Protein of unknown function VPA0982 | | Authors: | Tan, K, Duggan, E, Moy, S, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2007-07-05 | | Release date: | 2007-07-24 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.48 Å) | | Cite: | The crystal structure of a protein of unknown function, VPA0982 from Vibrio parahaemolyticus RIMD 2210633.
To be Published
|
|
2QSI
 
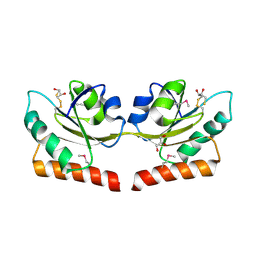 | | Crystal structure of putative hydrogenase expression/formation protein hupG from Rhodopseudomonas palustris CGA009 | | Descriptor: | 2,3-DIHYDROXY-1,4-DITHIOBUTANE, GLYCEROL, Putative hydrogenase expression/formation protein hupG | | Authors: | Nocek, B, Skarina, T, Kagan, O, Savchenko, A, Edwards, A, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2007-07-31 | | Release date: | 2007-08-14 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structure of putative hydrogenase expression/formation protein hupG from Rhodopseudomonas palustris CGA009.
To be Published
|
|
2QZ7
 
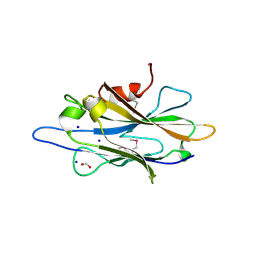 | | The crystal structure of a homologue of telluride resistance protein (TerD), SCO6318 from Streptomyces coelicolor A3(2) | | Descriptor: | 1,2-ETHANEDIOL, SODIUM ION, Uncharacterized protein SCO6318 | | Authors: | Tan, K, Xu, X, Zheng, Z, Savchenko, A, Edwards, A, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2007-08-16 | | Release date: | 2007-08-28 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | The crystal structure of a homologue of telluride resistance protein (TerD), SCO6318 from Streptomyces coelicolor A3(2).
To be Published
|
|
1ZKE
 
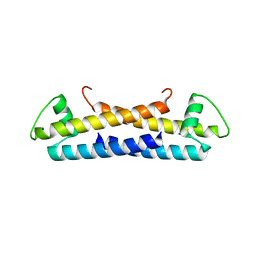 | | 1.6 A Crystal Structure of a Protein HP1531 of Unknown Function from Helicobacter pylori | | Descriptor: | Hypothetical protein HP1531, MAGNESIUM ION | | Authors: | Zhang, R, Skarina, T, Savchenko, A, Edwards, A, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2005-05-02 | | Release date: | 2005-06-28 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | 1.6A crystal structure of a hypothetical protein HP1531 from Helicobacter pylori 26695
To be Published
|
|
2R5S
 
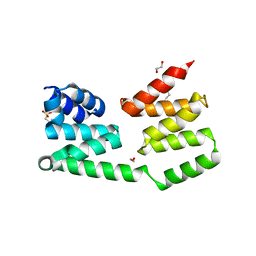 | | The crystal structure of a domain of protein VP0806 (unknown function) from Vibrio parahaemolyticus RIMD 2210633 | | Descriptor: | 1,2-ETHANEDIOL, 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, ACETATE ION, ... | | Authors: | Tan, K, Wu, R, Abdullah, J, Freeman, L, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2007-09-04 | | Release date: | 2007-09-18 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.14 Å) | | Cite: | The crystal structure of a domain of protein VP0806 (unknown function) from Vibrio parahaemolyticus RIMD 2210633.
To be Published
|
|
2AZ4
 
 | | Crystal Structure of a Protein of Unknown Function from Enterococcus faecalis V583 | | Descriptor: | ZINC ION, hypothetical protein EF2904 | | Authors: | Zhang, R, Maltseva, N, Moy, S, Collart, F, Cymborowski, M, Minor, W, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2005-09-09 | | Release date: | 2005-10-25 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | The 2.0 A crystal structure of a hypothetical protein from Enterococcus faecalis V583
To be Published
|
|
2QQZ
 
 | | Crystal structure of putative glyoxalase family protein from Bacillus anthracis | | Descriptor: | GLYCEROL, Glyoxalase family protein, putative, ... | | Authors: | Kim, Y, Joachimiak, G, Wu, R, Patterson, S, Gornicki, P, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2007-07-27 | | Release date: | 2007-08-14 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.92 Å) | | Cite: | Crystal Structure of Putative Glyoxalase Family Protein from Bacillus anthracis.
To be Published
|
|
2QX2
 
 | | Structure of the C-terminal domain of sex pheromone staph-cAM373 precursor from Staphylococcus aureus | | Descriptor: | 1,2-ETHANEDIOL, Sex pheromone staph-cAM373 | | Authors: | Cuff, M.E, Mussar, K, Hatzos, C, Gu, M, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2007-08-10 | | Release date: | 2007-09-11 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structure of the C-terminal domain of sex pheromone staph-cAM373 precursor from Staphylococcus aureus.
TO BE PUBLISHED
|
|
3R89
 
 | |
2PPX
 
 | | Crystal structure of a HTH XRE-family like protein from Agrobacterium tumefaciens | | Descriptor: | GLYCEROL, SULFATE ION, Uncharacterized protein Atu1735 | | Authors: | Cuff, M.E, Skarina, T, Onopriyenko, O, Edwards, A, Savchenko, A, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2007-04-30 | | Release date: | 2007-05-29 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structure of a HTH XRE-family like protein from Agrobacterium tumefaciens.
TO BE PUBLISHED
|
|
1TLJ
 
 | | Crystal Structure of Conserved Protein of Unknown Function SSO0622 from Sulfolobus solfataricus | | Descriptor: | Hypothetical UPF0130 protein SSO0622, SULFATE ION | | Authors: | Jia, Z, Wong, A.H.Y, Kudrytska, M, Skarina, T, Walker, J, Savchenko, A, Edwards, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2004-06-09 | | Release date: | 2004-08-03 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structural and functional characterization of the TYW3/Taw3 class of SAM-dependent methyltransferases.
Rna, 23, 2017
|
|
2P0S
 
 | | Structural Genomics, the crystal structure of a putative ABC transporter domain from Porphyromonas gingivalis W83 | | Descriptor: | ABC transporter, permease protein, putative, ... | | Authors: | Tan, K, Duggan, E, Abdullah, J, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2007-03-01 | | Release date: | 2007-04-03 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | The crystal structure of a putative ABC transporter domain from Porphyromonas gingivalis W83
To be Published
|
|
