6D8R
 
 | |
6D8U
 
 | |
6D9P
 
 | |
6D8Y
 
 | |
6D93
 
 | |
6D3T
 
 | |
6D8S
 
 | |
1QYG
 
 | | ANTI-COCAINE ANTIBODY M82G2 COMPLEXED WITH BENZOYLECGONINE | | Descriptor: | 3-(BENZOYLOXY)-8-METHYL-8-AZABICYCLO[3.2.1]OCTANE-2-CARBOXYLIC ACID, FAB M82G2, HEAVY CHAIN, ... | | Authors: | Pozharski, E, Hewagama, A, Shanafelt, A, Petsko, G, Ringe, D. | | Deposit date: | 2003-09-10 | | Release date: | 2003-09-23 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.81 Å) | | Cite: | Diversity in hapten recognition: structural study of an anti-cocaine antibody M82G2.
J.Mol.Biol., 349, 2005
|
|
5XHI
 
 | | Crystal structure of Frog M-ferritin D38A mutant | | Descriptor: | CHLORIDE ION, Ferritin, middle subunit, ... | | Authors: | Jagdev, M.K, Vasudevan, D. | | Deposit date: | 2017-04-21 | | Release date: | 2017-08-09 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.26 Å) | | Cite: | Surface charge dependent separation of modified and hybrid ferritin in native PAGE: Impact of lysine 104
Biochim. Biophys. Acta, 1865, 2017
|
|
5XHO
 
 | | Crystal structure of Frog M-ferritin E135K mutant | | Descriptor: | CHLORIDE ION, Ferritin, middle subunit, ... | | Authors: | Jagdev, M.K, Vasudevan, D. | | Deposit date: | 2017-04-21 | | Release date: | 2017-08-09 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.73 Å) | | Cite: | Surface charge dependent separation of modified and hybrid ferritin in native PAGE: Impact of lysine 104
Biochim. Biophys. Acta, 1865, 2017
|
|
6EIA
 
 | | PepTSt in complex with HEPES (100 mM) | | Descriptor: | (2R)-2,3-DIHYDROXYPROPYL(7Z)-PENTADEC-7-ENOATE, (2S)-2,3-DIHYDROXYPROPYL(7Z)-PENTADEC-7-ENOATE, 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, ... | | Authors: | Martinez Molledo, M, Quistgaard, E.M, Loew, C. | | Deposit date: | 2017-09-18 | | Release date: | 2018-02-21 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Multispecific Substrate Recognition in a Proton-Dependent Oligopeptide Transporter.
Structure, 26, 2018
|
|
5AIU
 
 | | A complex of RNF4-RING domain, Ubc13-Ub (isopeptide crosslink) | | Descriptor: | 1,2-ETHANEDIOL, E3 UBIQUITIN-PROTEIN LIGASE RNF4, POLYUBIQUITIN-C, ... | | Authors: | Branigan, E, Naismith, J.H. | | Deposit date: | 2015-02-17 | | Release date: | 2015-07-08 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.21 Å) | | Cite: | Structural Basis for the Ring Catalyzed Synthesis of K63 Linked Ubiquitin Chains
Nat.Struct.Mol.Biol., 22, 2015
|
|
1C2X
 
 | |
4HVI
 
 | |
5AIT
 
 | | A complex of of RNF4-RING domain, UbeV2, Ubc13-Ub (isopeptide crosslink) | | Descriptor: | E3 UBIQUITIN-PROTEIN LIGASE RNF4, POLYUBIQUITIN-C, UBIQUITIN-CONJUGATING ENZYME E2 N, ... | | Authors: | Branigan, E, Naismith, J.H. | | Deposit date: | 2015-02-17 | | Release date: | 2015-07-08 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (3.4 Å) | | Cite: | Structural Basis for the Ring Catalyzed Synthesis of K63 Linked Ubiquitin Chains
Nat.Struct.Mol.Biol., 22, 2015
|
|
4HVG
 
 | |
5LQS
 
 | | Structure of quinolinate synthase Y21F mutant in complex with substrate-derived quinolinate | | Descriptor: | CHLORIDE ION, IRON/SULFUR CLUSTER, QUINOLINIC ACID, ... | | Authors: | Volbeda, A, Fontecilla-Camps, J.C. | | Deposit date: | 2016-08-17 | | Release date: | 2016-08-31 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal Structures of Quinolinate Synthase in Complex with a Substrate Analogue, the Condensation Intermediate, and Substrate-Derived Product.
J.Am.Chem.Soc., 138, 2016
|
|
4HVH
 
 | |
7U0P
 
 | | SARS-Cov2 S protein structure in complex with neutralizing monoclonal antibody 002-S21F2 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Spike glycoprotein, ... | | Authors: | Patel, A, Ortlund, E. | | Deposit date: | 2022-02-18 | | Release date: | 2022-08-10 | | Last modified: | 2023-02-22 | | Method: | ELECTRON MICROSCOPY (3.76 Å) | | Cite: | Structural insights for neutralization of Omicron variants BA.1, BA.2, BA.4, and BA.5 by a broadly neutralizing SARS-CoV-2 antibody.
Sci Adv, 8, 2022
|
|
4HVD
 
 | | JAK3 kinase domain in complex with 2-Cyclopropyl-5H-pyrrolo[2,3-b]pyrazine-7-carboxylic acid ((S)-1,2,2-trimethyl-propyl)-amide | | Descriptor: | 1-phenylurea, 2-cyclopropyl-N-[(2S)-3,3-dimethylbutan-2-yl]-5H-pyrrolo[2,3-b]pyrazine-7-carboxamide, Tyrosine-protein kinase JAK3 | | Authors: | Kuglstatter, A, Shao, A. | | Deposit date: | 2012-11-06 | | Release date: | 2013-01-16 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | 3-Amido Pyrrolopyrazine JAK Kinase Inhibitors: Development of a JAK3 vs JAK1 Selective Inhibitor and Evaluation in Cellular and in Vivo Models.
J.Med.Chem., 56, 2013
|
|
7UPL
 
 | | SARS-Cov2 Omicron varient S protein structure in complex with neutralizing monoclonal antibody 002-S21F2 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Spike glycoprotein, ... | | Authors: | Patel, A, Ortlund, E. | | Deposit date: | 2022-04-15 | | Release date: | 2022-08-10 | | Last modified: | 2023-02-22 | | Method: | ELECTRON MICROSCOPY (4.1 Å) | | Cite: | Structural insights for neutralization of Omicron variants BA.1, BA.2, BA.4, and BA.5 by a broadly neutralizing SARS-CoV-2 antibody.
Sci Adv, 8, 2022
|
|
7UOW
 
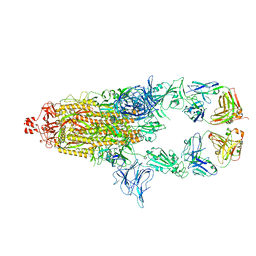 | | SARS-Cov2 S protein structure in complex with neutralizing monoclonal antibody 034_32 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Monoclonal antibody 034_32 heavy chain, ... | | Authors: | Patel, A, Ortlund, E. | | Deposit date: | 2022-04-14 | | Release date: | 2023-04-19 | | Last modified: | 2023-09-13 | | Method: | ELECTRON MICROSCOPY (4.4 Å) | | Cite: | Molecular basis of SARS-CoV-2 Omicron variant evasion from shared neutralizing antibody response.
Structure, 31, 2023
|
|
8W00
 
 | |
8VZX
 
 | |
8PKE
 
 | |
