8P5Y
 
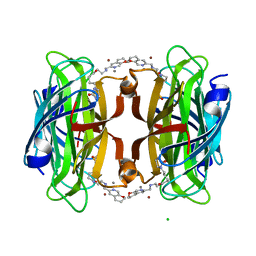 | | Artificial transfer hydrogenase with a Mn-12 cofactor and Streptavidin S112Y-K121M mutant | | Descriptor: | 5-[(3~{a}~{S},4~{S},6~{a}~{R})-2-oxidanylidene-1,3,3~{a},4,6,6~{a}-hexahydrothieno[3,4-d]imidazol-4-yl]-~{N}-[2-(3,4-dihydro-2~{H}-pyrano[2,3-c]pyridin-6-ylmethylamino)ethyl]pentanamide, BROMIDE ION, CHLORIDE ION, ... | | Authors: | Lau, K, Wang, W, Pojer, F, Larabi, A. | | Deposit date: | 2023-05-24 | | Release date: | 2023-09-13 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.88 Å) | | Cite: | Manganese Transfer Hydrogenases Based on the Biotin-Streptavidin Technology.
Angew.Chem.Int.Ed.Engl., 62, 2023
|
|
8P5Z
 
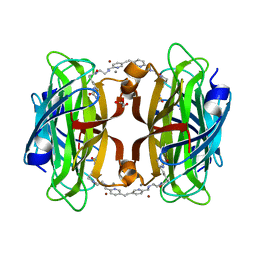 | | Artificial transfer hydrogenase with a Mn-5 cofactor and Streptavidin S112Y-K121M mutant | | Descriptor: | 5-[(3~{a}~{S},4~{S},6~{a}~{R})-2-oxidanylidene-1,3,3~{a},4,6,6~{a}-hexahydrothieno[3,4-d]imidazol-4-yl]-~{N}-[2-[(5-methylpyridin-2-yl)methylamino]ethyl]pentanamide, BROMIDE ION, GLYCEROL, ... | | Authors: | Lau, K, Wang, W, Pojer, F, Larabi, A. | | Deposit date: | 2023-05-24 | | Release date: | 2023-09-13 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.56 Å) | | Cite: | Manganese Transfer Hydrogenases Based on the Biotin-Streptavidin Technology.
Angew.Chem.Int.Ed.Engl., 62, 2023
|
|
2VG1
 
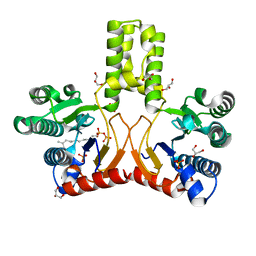 | | Rv1086 E,E-farnesyl diphosphate complex | | Descriptor: | FARNESYL DIPHOSPHATE, GLYCEROL, PHOSPHATE ION, ... | | Authors: | Naismith, J.H, Wang, W, Dong, C. | | Deposit date: | 2007-11-07 | | Release date: | 2007-11-13 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | The structural basis of chain length control in Rv1086.
J. Mol. Biol., 381, 2008
|
|
2VG0
 
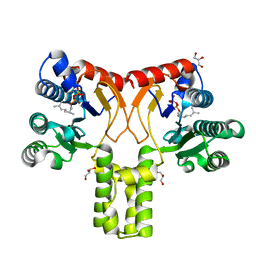 | | Rv1086 citronellyl pyrophosphate complex | | Descriptor: | GERANYL DIPHOSPHATE, GLYCEROL, SHORT-CHAIN Z-ISOPRENYL DIPHOSPHATE SYNTHETASE | | Authors: | Naismith, J.H, Wang, W, Dong, C. | | Deposit date: | 2007-11-07 | | Release date: | 2007-11-13 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | The structural basis of chain length control in Rv1086.
J. Mol. Biol., 381, 2008
|
|
2VFW
 
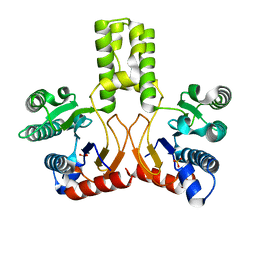 | | Rv1086 native | | Descriptor: | SHORT-CHAIN Z-ISOPRENYL DIPHOSPHATE SYNTHETASE, SULFATE ION | | Authors: | Naismith, J.H, Wang, W, Dong, C. | | Deposit date: | 2007-11-05 | | Release date: | 2007-11-13 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | The structural basis of chain length control in Rv1086.
J. Mol. Biol., 381, 2008
|
|
2VG2
 
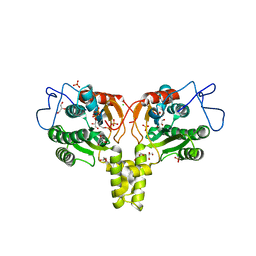 | | Rv2361 with IPP | | Descriptor: | 3-METHYLBUT-3-ENYL TRIHYDROGEN DIPHOSPHATE, CHLORIDE ION, DIPHOSPHATE, ... | | Authors: | Naismith, J.H, Wang, W, Dong, C. | | Deposit date: | 2007-11-07 | | Release date: | 2007-11-13 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | The structural basis of chain length control in Rv1086.
J. Mol. Biol., 381, 2008
|
|
2VG3
 
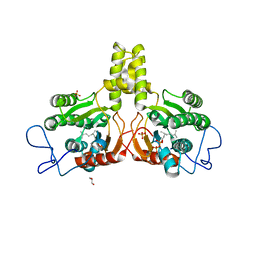 | | Rv2361 with citronellyl pyrophosphate | | Descriptor: | CHLORIDE ION, GERANYL DIPHOSPHATE, GLYCEROL, ... | | Authors: | Naismith, J.H, Wang, W, Dong, C. | | Deposit date: | 2007-11-08 | | Release date: | 2008-05-06 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | The structural basis of chain length control in Rv1086.
J. Mol. Biol., 381, 2008
|
|
2VG4
 
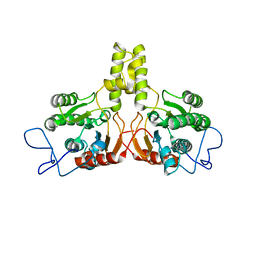 | | Rv2361 native | | Descriptor: | UNDECAPRENYL PYROPHOSPHATE SYNTHETASE | | Authors: | Naismith, J.H, Wang, W, Dong, C. | | Deposit date: | 2007-11-08 | | Release date: | 2007-11-27 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | The structural basis of chain length control in Rv1086.
J. Mol. Biol., 381, 2008
|
|
2XUV
 
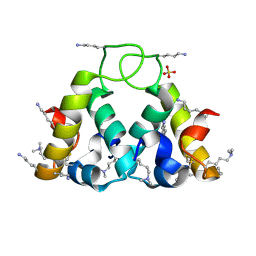 | | The structure of HdeB | | Descriptor: | HDEB, SULFATE ION | | Authors: | Naismith, J.H, Wang, W. | | Deposit date: | 2010-10-21 | | Release date: | 2011-08-24 | | Last modified: | 2012-01-25 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Salt Bridges Regulate Both Dimer Formation and Monomeric Flexibility in Hdeb and May Have a Role in Periplasmic Chaperone Function.
J.Mol.Biol., 415, 2012
|
|
5GTB
 
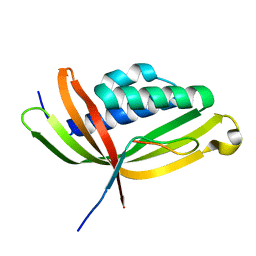 | |
4ZMH
 
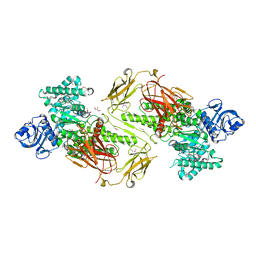 | | Crystal structure of a five-domain GH115 alpha-Glucuronidase from the Marine Bacterium Saccharophagus degradans 2-40T | | Descriptor: | ACETATE ION, GLYCEROL, PHOSPHATE ION, ... | | Authors: | Nocek, B, Cui, H, Wang, W, Savchenko, A. | | Deposit date: | 2015-05-04 | | Release date: | 2016-05-04 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.93 Å) | | Cite: | Biochemical and Structural Characterization of a Five-domain GH115 alpha-Glucuronidase from the Marine Bacterium Saccharophagus degradans 2-40T.
J.Biol.Chem., 291, 2016
|
|
4I6P
 
 | | Crystal structure of Par3-NTD domain | | Descriptor: | Partitioning defective 3 homolog | | Authors: | Wang, W, Gao, F, Gong, W, Sun, F, Feng, W. | | Deposit date: | 2012-11-29 | | Release date: | 2013-07-17 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Structural insights into the intrinsic self-assembly of par-3 N-terminal domain.
Structure, 21, 2013
|
|
3HVL
 
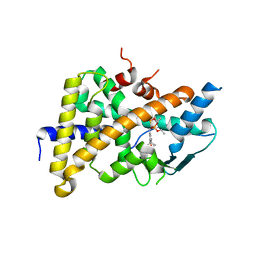 | | Tethered PXR-LBD/SRC-1p complexed with SR-12813 | | Descriptor: | Pregnane X receptor, Linker, Steroid receptor coactivator 1, ... | | Authors: | Lesburg, C.A, Wang, W, Prosise, W.W, Chen, J, Taremi, S.S, Le, H.V, Madison, V, Cui, X, Thomas, A, Cheng, K.C. | | Deposit date: | 2009-06-16 | | Release date: | 2009-08-04 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Construction and characterization of a fully active PXR/SRC-1 tethered protein with increased stability
Protein Eng.Des.Sel., 21, 2008
|
|
3R1G
 
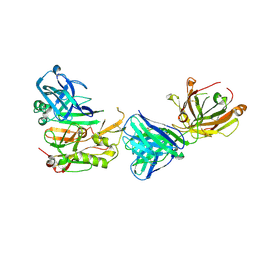 | | Structure Basis of Allosteric Inhibition of BACE1 by an Exosite-Binding Antibody | | Descriptor: | Beta-secretase 1, FAB of YW412.8.31 antibody heavy chain, FAB of YW412.8.31 antibody light chain | | Authors: | Wang, W, Rouge, L, Wu, P, Chiu, C, Chen, Y, Wu, Y, Watts, R.J. | | Deposit date: | 2011-03-10 | | Release date: | 2011-06-08 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | A Therapeutic Antibody Targeting BACE1 Inhibits Amyloid-{beta} Production in Vivo.
Sci Transl Med, 3, 2011
|
|
3NOJ
 
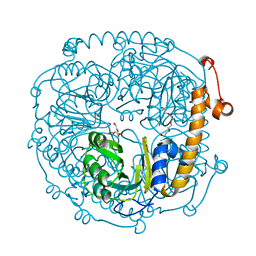 | | The structure of HMG/CHA aldolase from the protocatechuate degradation pathway of Pseudomonas putida | | Descriptor: | 4-carboxy-4-hydroxy-2-oxoadipate aldolase/oxaloacetate decarboxylase, MAGNESIUM ION, PYRUVIC ACID, ... | | Authors: | Kimber, M.S, Wang, W, Mazurkewich, S, Seah, S.Y.K. | | Deposit date: | 2010-06-25 | | Release date: | 2010-09-15 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.82 Å) | | Cite: | Structural and Kinetic Characterization of 4-Hydroxy-4-methyl-2-oxoglutarate/4-Carboxy-4-hydroxy-2-oxoadipate Aldolase, a Protocatechuate Degradation Enzyme Evolutionarily Convergent with the HpaI and DmpG Pyruvate Aldolases.
J.Biol.Chem., 285, 2010
|
|
8GYN
 
 | | zebrafish TIPE1 strucutre in complex with PE | | Descriptor: | Tumor necrosis factor alpha-induced protein 8-like protein 1, [(2~{R})-1-[2-azanylethoxy(oxidanyl)phosphoryl]oxy-3-hexadecanoyloxy-propan-2-yl] (~{Z})-octadec-9-enoate | | Authors: | Wang, W, Cao, S.J. | | Deposit date: | 2022-09-23 | | Release date: | 2023-04-19 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.38 Å) | | Cite: | Structural insight into TIPE1 functioning as a lipid transfer protein.
J.Biomol.Struct.Dyn., 41, 2023
|
|
7XUR
 
 | |
4I35
 
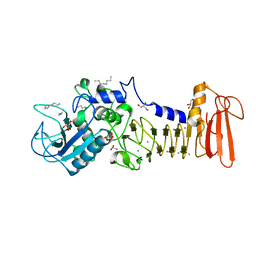 | | The crystal structure of serralysin | | Descriptor: | CALCIUM ION, GLYCEROL, HEXANE, ... | | Authors: | Zou, M, Ran, T, Xu, D, Wang, W. | | Deposit date: | 2012-11-24 | | Release date: | 2013-11-27 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.501 Å) | | Cite: | The crystal structure of serralysin
To be Published
|
|
3SV5
 
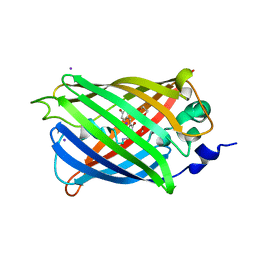 | | Engineered medium-affinity halide-binding protein derived from YFP: iodide complex | | Descriptor: | 1,2-ETHANEDIOL, FORMIC ACID, Green fluorescent protein, ... | | Authors: | Wang, W, Grimley, J.S, Beese, L.S, Hellinga, H.W. | | Deposit date: | 2011-07-12 | | Release date: | 2012-07-18 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.53 Å) | | Cite: | Visualization of Synaptic Inhibition with an Optogenetic Sensor Developed by Cell-Free Protein Engineering Automation.
J.Neurosci., 33, 2013
|
|
3ST0
 
 | | Engineered medium-affinity halide-binding protein derived from YFP: halide-free | | Descriptor: | 1,2-ETHANEDIOL, FORMIC ACID, Green fluorescent protein | | Authors: | Wang, W, Grimley, J.S, Beese, L.S, Hellinga, H.W. | | Deposit date: | 2011-07-08 | | Release date: | 2012-07-11 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.19 Å) | | Cite: | Visualization of Synaptic Inhibition with an Optogenetic Sensor Developed by Cell-Free Protein Engineering Automation.
J.Neurosci., 33, 2013
|
|
4O14
 
 | | Structural Basis for Resistance to Diverse Classes of NAMPT Inhibitors | | Descriptor: | 1,2-ETHANEDIOL, Nicotinamide phosphoribosyltransferase, PHOSPHATE ION | | Authors: | Oh, A, Coons, M, Brillantes, B, Wang, W. | | Deposit date: | 2013-12-15 | | Release date: | 2014-10-22 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.871 Å) | | Cite: | Structural Basis for Resistance to Diverse Classes of NAMPT Inhibitors.
Plos One, 9, 2014
|
|
4O1C
 
 | | The crystal structures of a mutant NAMPT H191R | | Descriptor: | 1,2-ETHANEDIOL, Nicotinamide phosphoribosyltransferase, PHOSPHATE ION | | Authors: | Oh, A, Coons, M, Brillantes, B, Wang, W. | | Deposit date: | 2013-12-15 | | Release date: | 2014-10-22 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.092 Å) | | Cite: | Structural Basis for Resistance to Diverse Classes of NAMPT Inhibitors.
Plos One, 9, 2014
|
|
4O15
 
 | | The crystal structure of a mutant NAMPT (S165F) in complex with GNE-618 | | Descriptor: | 1,2-ETHANEDIOL, N-(4-{[3-(trifluoromethyl)phenyl]sulfonyl}benzyl)-2H-pyrazolo[3,4-b]pyridine-5-carboxamide, Nicotinamide phosphoribosyltransferase, ... | | Authors: | Oh, A, Coons, M, Brillantes, B, Wang, W. | | Deposit date: | 2013-12-15 | | Release date: | 2014-10-22 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structural Basis for Resistance to Diverse Classes of NAMPT Inhibitors.
Plos One, 9, 2014
|
|
4O16
 
 | | Structural Basis for Resistance to Diverse Classes of NAMPT Inhibitors | | Descriptor: | 1,2-ETHANEDIOL, 6-({4-[(3,5-difluorophenyl)sulfonyl]benzyl}carbamoyl)-1-(5-O-phosphono-beta-D-ribofuranosyl)imidazo[1,2-a]pyridin-1-ium, Nicotinamide phosphoribosyltransferase, ... | | Authors: | Oh, A, Coons, M, Brillantes, B, Wang, W. | | Deposit date: | 2013-12-15 | | Release date: | 2014-10-22 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.783 Å) | | Cite: | Structural Basis for Resistance to Diverse Classes of NAMPT Inhibitors.
Plos One, 9, 2014
|
|
4O19
 
 | | The crystal structure of a mutant NAMPT (G217V) | | Descriptor: | 1,2-ETHANEDIOL, Nicotinamide phosphoribosyltransferase, PHOSPHATE ION | | Authors: | Oh, A, Coons, M, Brillantes, B, Wang, W. | | Deposit date: | 2013-12-15 | | Release date: | 2014-10-22 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Structural Basis for Resistance to Diverse Classes of NAMPT Inhibitors.
Plos One, 9, 2014
|
|
