6P6K
 
 | |
6P7Z
 
 | |
6PAF
 
 | |
6VG0
 
 | | CRYSTAL STRUCTURE OF HUMAN CYTOSOLIC ISOCITRATE DEHYDROGENASE (IDH1) R132H MUTANT IN COMPLEX WITH NADPH and AGI-15056 | | Descriptor: | Isocitrate dehydrogenase [NADP] cytoplasmic, NADPH DIHYDRO-NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, N~2~,N~4~-bis[(1R)-1-cyclopropylethyl]-6-[6-(trifluoromethyl)pyridin-2-yl]-1,3,5-triazine-2,4-diamine | | Authors: | Padyana, A, Jin, L. | | Deposit date: | 2020-01-07 | | Release date: | 2020-02-05 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.66 Å) | | Cite: | Vorasidenib (AG-881): A First-in-Class, Brain-Penetrant Dual Inhibitor of Mutant IDH1 and 2 for Treatment of Glioma.
Acs Med.Chem.Lett., 11, 2020
|
|
6VEI
 
 | | Crystal Structure of Human Cytosolic Isocitrate Dehydrogenase (IDH1) R132H Mutant in Complex with NADPH and AG-881 (Vorasidenib) Inhibitor | | Descriptor: | 3,6,9,12,15,18,21,24,27,30,33,36-dodecaoxaoctatriacontane-1,38-diol, 6-(6-chloropyridin-2-yl)-N2,N4-bis[(2R)-1,1,1-trifluoropropan-2-yl]-1,3,5-triazine-2,4-diamine, ACETATE ION, ... | | Authors: | Padyana, A, Jin, L. | | Deposit date: | 2020-01-02 | | Release date: | 2020-02-05 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Vorasidenib (AG-881): A First-in-Class, Brain-Penetrant Dual Inhibitor of Mutant IDH1 and 2 for Treatment of Glioma.
Acs Med.Chem.Lett., 11, 2020
|
|
6VFZ
 
 | | Crystal Structure of Human Mitochondrial Isocitrate Dehydrogenase (IDH2) R140Q Mutant Homodimer in Complex with NADPH and AG-881 (Vorasidenib) Inhibitor. | | Descriptor: | 6-(6-chloropyridin-2-yl)-N2,N4-bis[(2R)-1,1,1-trifluoropropan-2-yl]-1,3,5-triazine-2,4-diamine, CALCIUM ION, Isocitrate dehydrogenase [NADP], ... | | Authors: | Padyana, A, Jin, L. | | Deposit date: | 2020-01-07 | | Release date: | 2020-02-05 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.99 Å) | | Cite: | Vorasidenib (AG-881): A First-in-Class, Brain-Penetrant Dual Inhibitor of Mutant IDH1 and 2 for Treatment of Glioma.
Acs Med.Chem.Lett., 11, 2020
|
|
3H98
 
 | | Crystal structure of HCV NS5b 1b with (1,1-dioxo-2H-[1,2,4]benzothiadiazin-3-yl) azolo[1,5-a]pyrimidine derivative | | Descriptor: | GLYCEROL, N-{3-[5-hydroxy-8-(3-methylbutyl)-7-oxo-7,8-dihydroimidazo[1,2-a]pyrimidin-6-yl]-1,1-dioxido-4H-1,2,4-benzothiadiazin-7-yl}methanesulfonamide, RNA-directed RNA polymerase | | Authors: | Wang, G, Lei, H, Wang, X, Das, D, Mackinnon, C, Montalbetti, C.A.G, Mears, R, Gai, X, Bailey, S, Ruhrmund, D, Hooi, L, Misialek, S, Rajagopalan, R, Cheng, R.K.Y, Barker, J.L, Felicetti, B, Stoycheva, A, Buckman, B, Kossen, K, Seiwert, S, Beigelmana, L. | | Deposit date: | 2009-04-30 | | Release date: | 2009-10-13 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | HCV NS5B polymerase inhibitors 2: Synthesis and in vitro activity of (1,1-dioxo-2H-[1,2,4]benzothiadiazin-3-yl) azolo[1,5-a]pyridine and azolo[1,5-a]pyrimidine derivatives.
Bioorg.Med.Chem.Lett., 19, 2009
|
|
4WXP
 
 | |
4RLP
 
 | | Human p70s6k1 with ruthenium-based inhibitor FL772 | | Descriptor: | CHLORIDE ION, [(amino-kappaN)methanethiolato](3-fluoro-9-methoxypyrido[2,3-a]pyrrolo[3,4-c]carbazole-5,7(6H,12H)-dionato-kappa~2~N,N')(N-methyl-1,4,7-trithiecan-9-amine-kappa~3~S~1~,S~4~,S~7~)ruthenium, p70S6K1 | | Authors: | Domsic, J.F, Barber-Rotenberg, J, Salami, J, Qin, J, Marmorstein, R. | | Deposit date: | 2014-10-17 | | Release date: | 2015-01-21 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.79 Å) | | Cite: | Development of Organometallic S6K1 Inhibitors.
J.Med.Chem., 58, 2015
|
|
5D7E
 
 | | Crystal structure of Taf14 YEATS domain in complex with H3K9ac | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, DI(HYDROXYETHYL)ETHER, H3K9ac, ... | | Authors: | Andrews, F.H, Shanle, E.K, Strahl, B.D, Kutateladze, T.G. | | Deposit date: | 2015-08-13 | | Release date: | 2015-09-23 | | Last modified: | 2019-12-11 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Association of Taf14 with acetylated histone H3 directs gene transcription and the DNA damage response.
Genes Dev., 29, 2015
|
|
4RLO
 
 | | Human p70s6k1 with ruthenium-based inhibitor EM5 | | Descriptor: | CHLORIDE ION, DIMETHYL SULFOXIDE, GLYCEROL, ... | | Authors: | Domsic, J.F, Barber-Rotenberg, J, Salami, J, Qin, J, Marmorstein, R. | | Deposit date: | 2014-10-17 | | Release date: | 2015-01-21 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.527 Å) | | Cite: | Development of Organometallic S6K1 Inhibitors.
J.Med.Chem., 58, 2015
|
|
5WJH
 
 | | Using sound pulses to solve the crystal harvesting bottleneck | | Descriptor: | (CARBAMOYLMETHYL-CARBOXYMETHYL-AMINO)-ACETIC ACID, CALCIUM ION, Proteinase K | | Authors: | Soares, A.S, Brennan, H.M, McCarthy, L, Leroy, L. | | Deposit date: | 2017-07-22 | | Release date: | 2017-08-02 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.63 Å) | | Cite: | Using sound pulses to solve the crystal-harvesting bottleneck.
Acta Crystallogr D Struct Biol, 74, 2018
|
|
2EIR
 
 | |
1BOZ
 
 | | STRUCTURE-BASED DESIGN AND SYNTHESIS OF LIPOPHILIC 2,4-DIAMINO-6-SUBSTITUTED QUINAZOLINES AND THEIR EVALUATION AS INHIBITORS OF DIHYDROFOLATE REDUCTASE AND POTENTIAL ANTITUMOR AGENTS | | Descriptor: | N6-(2,5-DIMETHOXY-BENZYL)-N6-METHYL-PYRIDO[2,3-D]PYRIMIDINE-2,4,6-TRIAMINE, NADPH DIHYDRO-NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, PROTEIN (DIHYDROFOLATE REDUCTASE) | | Authors: | Gangjee, A, Vidwans, A.P, Vasudevan, A, Queener, S.F, Kisliuk, R.L, Cody, V, Li, R, Galitsky, N, Luft, J.R, Pangborn, W. | | Deposit date: | 1998-08-06 | | Release date: | 1998-08-12 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structure-based design and synthesis of lipophilic 2,4-diamino-6-substituted quinazolines and their evaluation as inhibitors of dihydrofolate reductases and potential antitumor agents.
J.Med.Chem., 41, 1998
|
|
2EIO
 
 | |
2EIQ
 
 | |
6UGJ
 
 | |
6UGG
 
 | | Structure of unmodified E. coli tRNA(Asp) | | Descriptor: | tRNAasp | | Authors: | Chan, C.W, Mondragon, A. | | Deposit date: | 2019-09-26 | | Release date: | 2020-01-01 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Crystal structures of an unmodified bacterial tRNA reveal intrinsic structural flexibility and plasticity as general properties of unbound tRNAs.
Rna, 26, 2020
|
|
6UGI
 
 | |
6WNS
 
 | |
6WN0
 
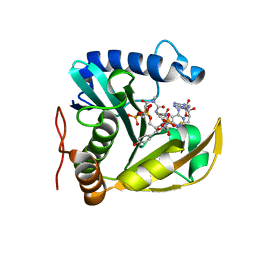 | | The structure of a CoA-dependent acyl-homoserine lactone synthase, RpaI, with the adduct of SAH and p-coumaroyl CoA | | Descriptor: | (2S)-4-({[(2S,3S,4R,5R)-5-(6-amino-9H-purin-9-yl)-3,4-dihydroxytetrahydrofuran-2-yl]methyl}sulfanyl)-2-{[(2E)-3-(cis-4-hydroxycyclohexa-2,5-dien-1-yl)prop-2-enoyl]amino}butanoic acid, 4-coumaroyl-homoserine lactone synthase, COENZYME A | | Authors: | Dong, S.-H, Nair, S.K. | | Deposit date: | 2020-04-22 | | Release date: | 2020-05-13 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Structure-Guided Biochemical Analysis of Quorum Signal Synthase Specificities.
Acs Chem.Biol., 15, 2020
|
|
6HGY
 
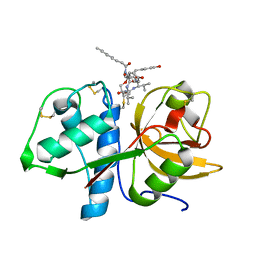 | | CRYSTAL STRUCTURE OF CATHEPSIN K WITH N-DESMETHYL THALASSOSPIRAMIDE C | | Descriptor: | Cathepsin K, THALASSOSPIRAMIDE C | | Authors: | Zakarian, A, Buckman, B.O, Adler, M, Griessner, A, Blaesse, M. | | Deposit date: | 2018-08-23 | | Release date: | 2019-06-26 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Total Synthesis of Covalent Cysteine Protease Inhibitor N-Desmethyl Thalassospiramide C and Crystallographic Evidence for Its Mode of Action.
Org.Lett., 21, 2019
|
|
6BQK
 
 | |
6BQJ
 
 | |
3PHE
 
 | | HCV NS5B with a bound quinolone inhibitor | | Descriptor: | 4-chlorobenzyl 6-fluoro-7-(4-methylpiperazin-1-yl)-1-[4-(methylsulfonyl)benzyl]-4-oxo-1,4-dihydroquinoline-3-carboxylate, HCV encoded nonstructural 5B protein | | Authors: | Somoza, J.R, To, N, Lehoux, I. | | Deposit date: | 2010-11-03 | | Release date: | 2010-11-17 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Quinolones as HCV NS5B polymerase inhibitors.
Bioorg.Med.Chem.Lett., 21, 2011
|
|
