6RV1
 
 | | human Alanine:Glyoxylate Aminotransferase major allele (AGT-Ma) | | Descriptor: | PYRIDOXAL-5'-PHOSPHATE, Serine--pyruvate aminotransferase | | Authors: | Giardina, G, Cutruzzola, F, Cellini, B, Mirco, D. | | Deposit date: | 2019-05-30 | | Release date: | 2020-04-08 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Cycloserine enantiomers are reversible inhibitors of human alanine:glyoxylate aminotransferase: implications for Primary Hyperoxaluria type 1.
Biochem.J., 476, 2019
|
|
6E8R
 
 | |
6S8W
 
 | | Aromatic aminotransferase AroH (Aro8) form Aspergillus fumigatus in complex with PLP (internal aldimine) | | Descriptor: | Aromatic aminotransferase Aro8, putative, FORMIC ACID, ... | | Authors: | Giardina, G, Mirco, D, Spizzichino, S, Zelante, T, Cutruzzola, F, Romani, L, Cellini, B. | | Deposit date: | 2019-07-10 | | Release date: | 2020-08-26 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Crystal structure of Aspergillus fumigatus AroH, an aromatic amino acid aminotransferase
Proteins, 2021
|
|
2C3T
 
 | | Human glutathione-S-transferase T1-1, W234R mutant, apo form | | Descriptor: | GLUTATHIONE S-TRANSFERASE THETA 1 | | Authors: | Tars, K, Larsson, A.-K, Shokeer, A, Olin, B, Mannervik, B, Kleywegt, G.J. | | Deposit date: | 2005-10-12 | | Release date: | 2005-11-30 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structural Basis of the Suppressed Catalytic Activity of Wild-Type Human Glutathione Transferase T1-1 Compared to its W234R Mutant.
J.Mol.Biol., 355, 2006
|
|
2C3N
 
 | | Human glutathione-S-transferase T1-1, apo form | | Descriptor: | GLUTATHIONE S-TRANSFERASE THETA 1, IODIDE ION | | Authors: | Tars, K, Larsson, A.-K, Shokeer, A, Olin, B, Mannervik, B, Kleywegt, G.J. | | Deposit date: | 2005-10-11 | | Release date: | 2005-11-30 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Structural Basis of the Suppressed Catalytic Activity of Wild-Type Human Glutathione Transferase T1-1 Compared to its W234R Mutant.
J.Mol.Biol., 355, 2006
|
|
2C3Q
 
 | | Human glutathione-S-transferase T1-1 W234R mutant, complex with S- hexylglutathione | | Descriptor: | GLUTATHIONE S-TRANSFERASE THETA 1, IODIDE ION, S-HEXYLGLUTATHIONE | | Authors: | Tars, K, Larsson, A.-K, Shokeer, A, Olin, B, Mannervik, B, Kleywegt, G.J. | | Deposit date: | 2005-10-11 | | Release date: | 2005-11-30 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Structural Basis of the Suppressed Catalytic Activity of Wild-Type Human Glutathione Transferase T1-1 Compared to its W234R Mutant.
J.Mol.Biol., 355, 2006
|
|
1XE8
 
 | | Crystal structure of the YML079w protein from Saccharomyces cerevisiae reveals a new sequence family of the jelly roll fold. | | Descriptor: | ADENINE, CITRIC ACID, GLYCEROL, ... | | Authors: | Zhou, C.-Z, Meyer, P, Quevillon-Cheruel, S, Li de La Sierra-Gallay, I, Collinet, B, Graille, M, Leulliot, N, Sorel, I, Janin, J, Van Tilbeurgh, H. | | Deposit date: | 2004-09-09 | | Release date: | 2005-01-11 | | Last modified: | 2017-10-11 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Crystal structure of the YML079w protein from Saccharomyces cerevisiae reveals a new sequence family of the jelly-roll fold
Protein Sci., 14, 2005
|
|
8QUA
 
 | | GTP binding protein YsxC from Staphylococcus aureus | | Descriptor: | ACETYL GROUP, GLYCEROL, Probable GTP-binding protein EngB | | Authors: | Biktimirov, A, Islamov, D, Lazarenko, V, Fatkhullin, B, Validov, S, Yusupov, M, Usachev, K. | | Deposit date: | 2023-10-15 | | Release date: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structure of GTPase YsxC from Staphylococcus aureus.
Biochem.Biophys.Res.Commun., 699, 2024
|
|
1L3P
 
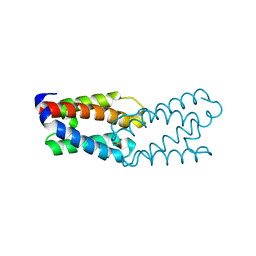 | | CRYSTAL STRUCTURE OF THE FUNCTIONAL DOMAIN OF THE MAJOR GRASS POLLEN ALLERGEN Phl p 5b | | Descriptor: | MAGNESIUM ION, PHOSPHATE ION, POLLEN ALLERGEN Phl p 5b | | Authors: | Rajashankar, K.R, Bufe, A, Weber, W, Eschenburg, S, Lindner, B, Betzel, C. | | Deposit date: | 2002-02-28 | | Release date: | 2003-02-28 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.98 Å) | | Cite: | Structure of the functional domain of the major grass-pollen allergen Phlp 5b.
Acta Crystallogr.,Sect.D, 58, 2002
|
|
1XE7
 
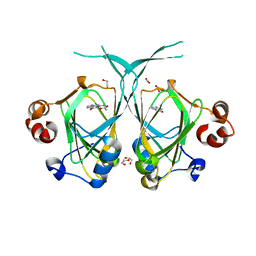 | | Crystal structure of the YML079w protein from Saccharomyces cerevisiae reveals a new sequence family of the jelly roll fold | | Descriptor: | 1,2-ETHANEDIOL, ACETIC ACID, GUANINE, ... | | Authors: | Zhou, C.-Z, Meyer, P, Quevillon-Cheruel, S, Li de La Sierra-Gallay, I, Collinet, B, Graille, M, Leulliot, N, Sorel, I, Janin, J, Van Tilbeurgh, H. | | Deposit date: | 2004-09-09 | | Release date: | 2005-01-11 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Crystal structure of the YML079w protein from Saccharomyces cerevisiae reveals a new sequence family of the jelly-roll fold
Protein Sci., 14, 2005
|
|
6BXH
 
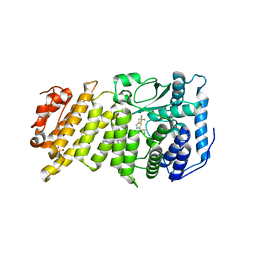 | | Menin in complex with MI-853 | | Descriptor: | 1-{2-[4-(fluoroacetyl)piperazin-1-yl]ethyl}-4-methyl-5-[(4-{[6-(2,2,2-trifluoroethyl)thieno[2,3-d]pyrimidin-4-yl]amino}piperidin-1-yl)methyl]-1H-indole-2-carbonitrile, DIMETHYL SULFOXIDE, Menin, ... | | Authors: | Borkin, D, Klossowski, S, Pollock, J, Linhares, B, Cierpicki, T, Grembecka, J. | | Deposit date: | 2017-12-18 | | Release date: | 2018-11-28 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.445 Å) | | Cite: | Menin in complex with MI-853
To Be Published
|
|
8TXY
 
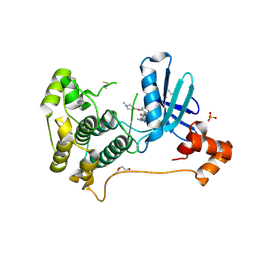 | | X-ray crystal structure of JRD-SIK1/2i-3 bound to a MARK2-SIK2 chimera | | Descriptor: | DI(HYDROXYETHYL)ETHER, N-[(5P,8R)-5-(2-cyano-5-{[(3R)-1-methylpyrrolidin-3-yl]methoxy}pyridin-4-yl)pyrazolo[1,5-a]pyridin-2-yl]cyclopropanecarboxamide, SULFATE ION, ... | | Authors: | Raymond, D.D, Lemke, C.T, Shaffer, P.L, Collins, B, Steele, R, Seierstad, M. | | Deposit date: | 2023-08-24 | | Release date: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Identification of highly selective SIK1/2 inhibitors that modulate innate immune activation and suppress intestinal inflammation.
Proc.Natl.Acad.Sci.USA, 121, 2024
|
|
5OFY
 
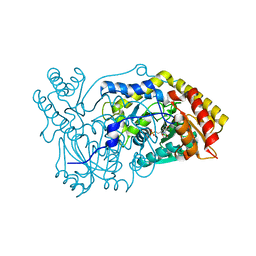 | | Crystal structure of the D183N variant of human Alanine:Glyoxylate Aminotransferase major allele (AGT-Ma) at pH 9.0. 2.8 Ang; internal aldimine with PLP in the active site | | Descriptor: | 1,4-DIETHYLENE DIOXIDE, PYRIDOXAL-5'-PHOSPHATE, Serine--pyruvate aminotransferase | | Authors: | Giardina, G, Cutruzzola, F, Borri Voltattorni, C, Cellini, B, Montioli, R. | | Deposit date: | 2017-07-11 | | Release date: | 2017-09-27 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Radiation damage at the active site of human alanine:glyoxylate aminotransferase reveals that the cofactor position is finely tuned during catalysis.
Sci Rep, 7, 2017
|
|
5OG0
 
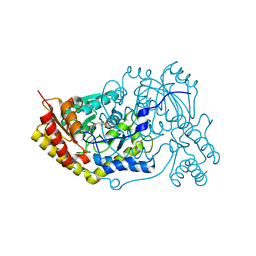 | | Crystal structure of human Alanine:Glyoxylate Aminotransferase major allele (AGT-Ma) at 2.5 Angstrom; internal aldimine with PLP in the active site | | Descriptor: | PYRIDOXAL-5'-PHOSPHATE, Serine--pyruvate aminotransferase | | Authors: | Giardina, G, Cutruzzola, F, Borri Voltattorni, C, Cellini, B, Montioli, R. | | Deposit date: | 2017-07-11 | | Release date: | 2017-09-27 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Radiation damage at the active site of human alanine:glyoxylate aminotransferase reveals that the cofactor position is finely tuned during catalysis.
Sci Rep, 7, 2017
|
|
3PSN
 
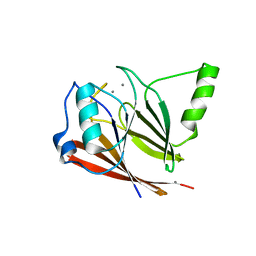 | | Crystal structure of mouse VPS29 complexed with Mn2+ | | Descriptor: | MANGANESE (II) ION, Vacuolar protein sorting-associated protein 29 | | Authors: | Swarbrick, J, Shaw, D, Chhabra, S, Ghai, R, Valkov, E, Norwood, S, Collins, B. | | Deposit date: | 2010-12-02 | | Release date: | 2010-12-15 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Conformational dynamics and biomolecular interactions of VPS29 studied by NMR and X-ray crystallography
To be Published
|
|
3PSO
 
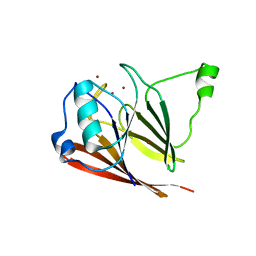 | | Crystal structure of mouse VPS29 complexed with Zn2+ | | Descriptor: | Vacuolar protein sorting-associated protein 29, ZINC ION | | Authors: | Swarbrick, J, Shaw, D, Chhabra, S, Ghai, R, Valkov, E, Norwood, S, Collins, B. | | Deposit date: | 2010-12-02 | | Release date: | 2010-12-15 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Conformational dynamics and biomolecular interactions of VPS29 studied by NMR and X-ray crystallography
To be Published
|
|
4HV0
 
 | | Structure and Function of AvtR, a Novel Transcriptional Regulator from a Hyperthermophilic Archaeal Lipothrixvirus | | Descriptor: | AvtR | | Authors: | Peixeiro, N, Keller, J, Collinet, B, Leulliot, N, Campanacci, V, Cortez, D, Cambillau, C, Nitta, K.R, Vincentelli, R, Forterre, P, Prangishvili, D, Sezonov, G, van Tilbeurgh, H. | | Deposit date: | 2012-11-05 | | Release date: | 2012-11-21 | | Last modified: | 2012-12-26 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structure and Function of AvtR, a Novel Transcriptional Regulator from a Hyperthermophilic Archaeal Lipothrixvirus.
J.Virol., 87, 2013
|
|
4I8A
 
 | | Alanine-glyoxylate aminotransferase variant S187F | | Descriptor: | GLYCEROL, Serine-pyruvate aminotransferase | | Authors: | Fodor, K, Oppici, E, Williams, C, Cellini, B, Wilmanns, M. | | Deposit date: | 2012-12-03 | | Release date: | 2013-05-01 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Crystal structure of the S187F variant of human liver alanine: Aminotransferase associated with primary hyperoxaluria type I and its functional implications.
Proteins, 81, 2013
|
|
3RBF
 
 | | Crystal structure of Human aromatic L-amino acid decarboxylase (AADC) in the apo form | | Descriptor: | Aromatic-L-amino-acid decarboxylase, CHLORIDE ION, PYRIDOXAL-5'-PHOSPHATE | | Authors: | Giardina, G, Montioli, R, Gianni, S, Cellini, B, Paiardini, A, Borri Voltattorni, C, Cutruzzola, F. | | Deposit date: | 2011-03-29 | | Release date: | 2011-10-19 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Open conformation of human DOPA decarboxylase reveals the mechanism of PLP addition to Group II decarboxylases.
Proc.Natl.Acad.Sci.USA, 108, 2011
|
|
2VCV
 
 | |
2VCT
 
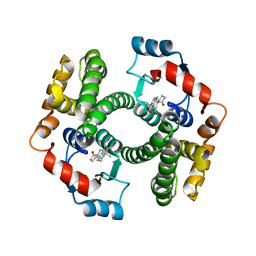 | |
1PKZ
 
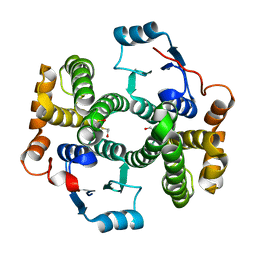 | | Crystal structure of human glutathione transferase (GST) A1-1 | | Descriptor: | 2-HYDROXYETHYL DISULFIDE, Glutathione S-transferase A1 | | Authors: | Grahn, E, Jakobsson, E, Gustafsson, A, Grehn, L, Olin, B, Wahlberg, M, Madsen, D, Kleywegt, G.J, Mannervik, B. | | Deposit date: | 2003-06-06 | | Release date: | 2004-06-22 | | Last modified: | 2018-03-07 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | New crystal structures of human glutathione transferase A1-1 shed light on glutathione binding and the conformation of the C-terminal helix.
Acta Crystallogr.,Sect.D, 62, 2006
|
|
1PL2
 
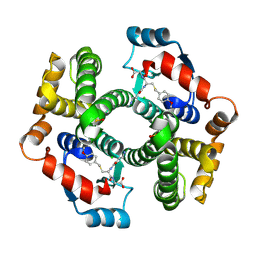 | | Crystal structure of human glutathione transferase (GST) A1-1 T68E mutant in complex with decarboxy-glutathione | | Descriptor: | CHLORIDE ION, Glutathione S-transferase A1, N-(4-AMINOBUTANOYL)-S-(4-METHOXYBENZYL)-L-CYSTEINYLGLYCINE | | Authors: | Grahn, E, Jakobsson, E, Gustafsson, A, Grehn, L, Olin, B, Wahlberg, M, Madsen, D, Kleywegt, G.J, Mannervik, B. | | Deposit date: | 2003-06-06 | | Release date: | 2004-06-22 | | Last modified: | 2021-10-27 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | New crystal structures of human glutathione transferase A1-1 shed light on glutathione binding and the conformation of the C-terminal helix.
Acta Crystallogr.,Sect.D, 62, 2006
|
|
1PKW
 
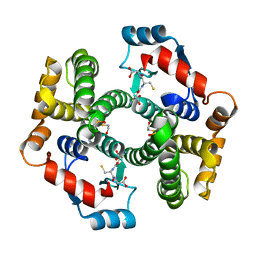 | | Crystal structure of human glutathione transferase (GST) A1-1 in complex with glutathione | | Descriptor: | 2-HYDROXYETHYL DISULFIDE, GLUTATHIONE, Glutathione S-transferase A1 | | Authors: | Grahn, E, Jakobsson, E, Gustafsson, A, Grehn, L, Olin, B, Wahlberg, M, Madsen, D, Kleywegt, G.J, Mannervik, B. | | Deposit date: | 2003-06-06 | | Release date: | 2004-06-22 | | Last modified: | 2018-03-07 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | New crystal structures of human glutathione transferase A1-1 shed light on glutathione binding and the conformation of the C-terminal helix.
Acta Crystallogr.,Sect.D, 62, 2006
|
|
1PL1
 
 | | Crystal structure of human glutathione transferase (GST) A1-1 in complex with a decarboxy-glutathione | | Descriptor: | CHLORIDE ION, Glutathione S-transferase A1, N-(4-AMINOBUTANOYL)-S-(4-METHOXYBENZYL)-L-CYSTEINYLGLYCINE | | Authors: | Grahn, E, Jakobsson, E, Gustafsson, A, Grehn, L, Olin, B, Wahlberg, M, Madsen, D, Kleywegt, G.J, Mannervik, B. | | Deposit date: | 2003-06-06 | | Release date: | 2004-06-22 | | Last modified: | 2018-03-07 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | New crystal structures of human glutathione transferase A1-1 shed light on glutathione binding and the conformation of the C-terminal helix.
Acta Crystallogr.,Sect.D, 62, 2006
|
|
