3JUE
 
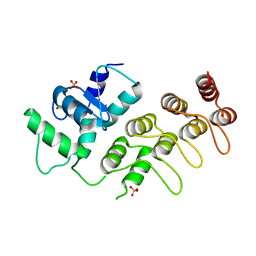 | | Crystal Structure of ArfGAP and ANK repeat domain of ACAP1 | | Descriptor: | ARFGAP with coiled-coil, ANK repeat and PH domain-containing protein 1, SULFATE ION, ... | | Authors: | Pang, X, Zhang, K, Ma, J, Zhou, Q, Sun, F. | | Deposit date: | 2009-09-15 | | Release date: | 2010-09-22 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Mechanistic insights into regulated cargo binding by ACAP1 protein
J.Biol.Chem., 287, 2012
|
|
3J17
 
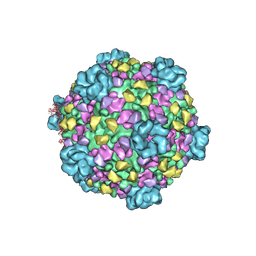 | | Structure of a transcribing cypovirus by cryo-electron microscopy | | Descriptor: | Structural protein VP3, Structural protein VP5, VP1 | | Authors: | Yang, C, Ji, G, Liu, H, Zhang, K, Liu, G, Sun, F, Zhu, P, Cheng, L. | | Deposit date: | 2011-12-25 | | Release date: | 2012-04-04 | | Last modified: | 2018-07-18 | | Method: | ELECTRON MICROSCOPY (4.1 Å) | | Cite: | Cryo-EM structure of a transcribing cypovirus.
Proc.Natl.Acad.Sci.USA, 109, 2012
|
|
3ZIF
 
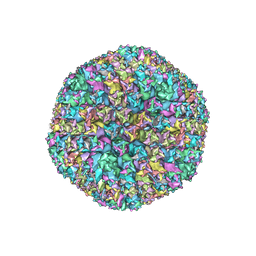 | | Cryo-EM structures of two intermediates provide insight into adenovirus assembly and disassembly | | Descriptor: | HEXON PROTEIN, PENTON PROTEIN, PIX, ... | | Authors: | Cheng, L, Huang, X, Li, X, Xiong, W, Sun, W, Yang, C, Zhang, K, Wang, Y, Liu, H, Ji, G, Sun, F, Zheng, C, Zhu, P. | | Deposit date: | 2013-01-09 | | Release date: | 2014-01-22 | | Last modified: | 2024-05-08 | | Method: | ELECTRON MICROSCOPY (4.5 Å) | | Cite: | Cryo-Em Structures of Two Bovine Adenovirus Type 3 Intermediates
Virology, 450, 2014
|
|
8XVB
 
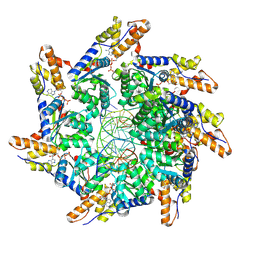 | | Cryo-EM structure of ATP-DNA-MuB filaments | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, ATP-dependent target DNA activator B, DNA (5'-D(P*AP*AP*AP*AP*AP*AP*AP*AP*AP*AP*AP*AP*AP*AP*AP*AP*AP*AP*AP*AP*AP*AP*AP*A)-3'), ... | | Authors: | Zhao, X, Zhang, K, Li, S. | | Deposit date: | 2024-01-14 | | Release date: | 2024-08-14 | | Last modified: | 2024-08-21 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Elucidating the Architectural dynamics of MuB filaments in bacteriophage Mu DNA transposition
Nat Commun, 15, 2024
|
|
7YTY
 
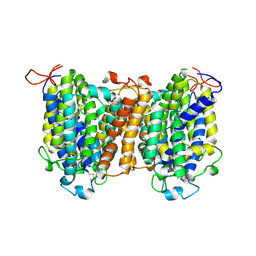 | | Mouse SVCT1 in an apo state | | Descriptor: | Solute carrier family 23 member 1 | | Authors: | She, J, Wang, M, He, J, Zhang, K, Li, S. | | Deposit date: | 2022-08-16 | | Release date: | 2023-05-03 | | Last modified: | 2024-10-09 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | Structural basis of vitamin C recognition and transport by mammalian SVCT1 transporter.
Nat Commun, 14, 2023
|
|
8XVC
 
 | | CryoEM structure of ADP-DNA-MuB conformation1 | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, ATP-dependent target DNA activator B | | Authors: | Zhao, X, Zhang, K, Li, S. | | Deposit date: | 2024-01-14 | | Release date: | 2024-08-14 | | Method: | ELECTRON MICROSCOPY (4.32 Å) | | Cite: | Elucidating the Architectural dynamics of MuB filaments in bacteriophage Mu DNA transposition.
Nat Commun, 15, 2024
|
|
7YTW
 
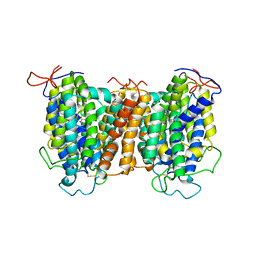 | | Structural basis of vitamin C recognition and transport by mammalian SVCT1 transporter | | Descriptor: | ASCORBIC ACID, SODIUM ION, Solute carrier family 23 member 1 | | Authors: | She, J, Wang, M, He, J, Zhang, K, Li, S. | | Deposit date: | 2022-08-16 | | Release date: | 2023-05-03 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Structural basis of vitamin C recognition and transport by mammalian SVCT1 transporter.
Nat Commun, 14, 2023
|
|
8XVD
 
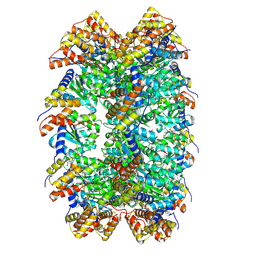 | | CryoEM structure of ADP-DNA-MuB conformation2 | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, ATP-dependent target DNA activator B | | Authors: | Zhao, X, Zhang, K, Li, S. | | Deposit date: | 2024-01-14 | | Release date: | 2024-08-14 | | Method: | ELECTRON MICROSCOPY (4.43 Å) | | Cite: | Elucidating the Architectural dynamics of MuB filaments in bacteriophage Mu DNA transposition.
Nat Commun, 15, 2024
|
|
7YPI
 
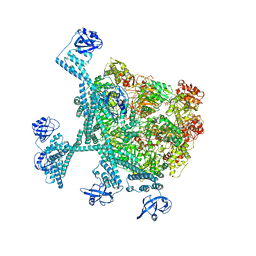 | | Spiral hexamer of the substrate-free Lon protease with a Y224S mutation | | Descriptor: | Lon protease, PHOSPHOTHIOPHOSPHORIC ACID-ADENYLATE ESTER | | Authors: | Li, S, Hsieh, K.Y, Kuo, C.I, Lee, S.H, Ho, M.R, Wang, C.H, Zhang, K, Chang, C.I. | | Deposit date: | 2022-08-03 | | Release date: | 2023-10-25 | | Last modified: | 2023-11-29 | | Method: | ELECTRON MICROSCOPY (3.8 Å) | | Cite: | A 5+1 assemble-to-activate mechanism of the Lon proteolytic machine.
Nat Commun, 14, 2023
|
|
7YPJ
 
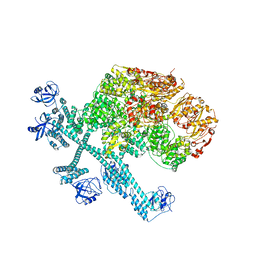 | | Spiral pentamer of the substrate-free Lon protease with a S678A mutation | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Lon protease | | Authors: | Li, S, Hsieh, K.Y, Kuo, C.I, Lee, S.H, Ho, M.R, Wang, C.H, Zhang, K, Chang, C.I. | | Deposit date: | 2022-08-03 | | Release date: | 2023-10-25 | | Last modified: | 2023-11-29 | | Method: | ELECTRON MICROSCOPY (3.8 Å) | | Cite: | A 5+1 assemble-to-activate mechanism of the Lon proteolytic machine.
Nat Commun, 14, 2023
|
|
7YPH
 
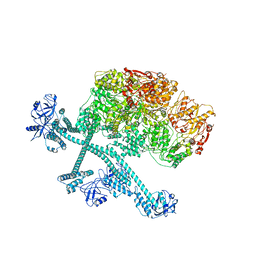 | | Open-spiral pentamer of the substrate-free Lon protease with a Y224S mutation | | Descriptor: | Lon protease, PHOSPHOTHIOPHOSPHORIC ACID-ADENYLATE ESTER | | Authors: | Li, S, Hsieh, K.Y, Kuo, C.I, Lee, S.H, Ho, M.R, Wang, C.H, Zhang, K, Chang, C.I. | | Deposit date: | 2022-08-03 | | Release date: | 2023-10-25 | | Last modified: | 2023-11-29 | | Method: | ELECTRON MICROSCOPY (3.68 Å) | | Cite: | A 5+1 assemble-to-activate mechanism of the Lon proteolytic machine.
Nat Commun, 14, 2023
|
|
7YPK
 
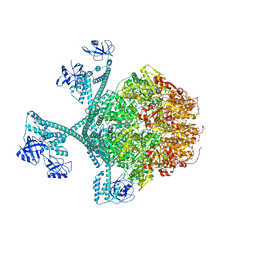 | | Close-ring hexamer of the substrate-bound Lon protease with an S678A mutation | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Lon protease, alpha-S1-casein | | Authors: | Li, S, Hsieh, K.Y, Kuo, C.I, Lee, S.H, Ho, M.R, Wang, C.H, Zhang, K, Chang, C.I. | | Deposit date: | 2022-08-03 | | Release date: | 2023-10-25 | | Last modified: | 2023-11-29 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | A 5+1 assemble-to-activate mechanism of the Lon proteolytic machine.
Nat Commun, 14, 2023
|
|
4MBQ
 
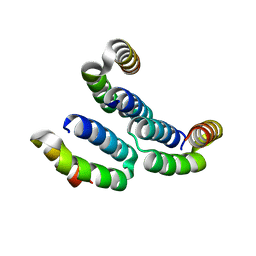 | | TPR3 of FimV from P. aeruginosa (PAO1) | | Descriptor: | Motility protein FimV | | Authors: | Nguyen, Y, Zhang, K, Daniel-Ivad, M, Robinson, H, Wolfram, F, Sugiman-Marangos, S.N, Junop, M.S, Burrows, L.L, Howell, P.L. | | Deposit date: | 2013-08-19 | | Release date: | 2014-08-20 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.006 Å) | | Cite: | Crystal structure of TPR2 from FimV
To be Published
|
|
4MAL
 
 | | TPR3 of FimV from P. aeruginosa (PAO1) | | Descriptor: | Motility protein FimV | | Authors: | Nguyen, Y, Zhang, K, Daniel-Ivad, M, Sugiman-Marangos, S.N, Junop, M.S, Burrows, L.L, Howell, P.L. | | Deposit date: | 2013-08-16 | | Release date: | 2014-08-20 | | Last modified: | 2016-02-24 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Crystal structure of TPR2 from FimV
To be Published
|
|
4NPH
 
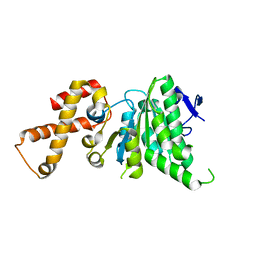 | | Crystal structure of SsaN from Salmonella enterica | | Descriptor: | Probable secretion system apparatus ATP synthase SsaN | | Authors: | Sugiman-Marangos, S.N, Zhang, K, Cooper, C.A, Coombes, B.K, Junop, M.S. | | Deposit date: | 2013-11-21 | | Release date: | 2014-07-23 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.09 Å) | | Cite: | Crystal structure of SsaN from Salmonella enterica
J.Biol.Chem., 2014
|
|
4OIY
 
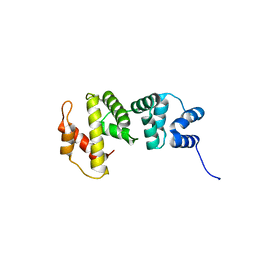 | | Crystal structure of Sec7p catalytic domain | | Descriptor: | MAGNESIUM ION, Protein transport protein SEC7 | | Authors: | Qiu, B, Zhang, K, Sun, F. | | Deposit date: | 2014-01-20 | | Release date: | 2014-07-02 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | C-terminal motif within Sec7 domain regulates guanine nucleotide exchange activity via tuning protein conformation
Biochem.Biophys.Res.Commun., 446, 2014
|
|
8WCN
 
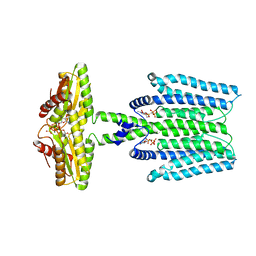 | | Cryo-EM structure of PAO1-ImcA with GMPCPP | | Descriptor: | Diguanylate cyclase, MAGNESIUM ION, PHOSPHOMETHYLPHOSPHONIC ACID GUANYLATE ESTER | | Authors: | Zhan, X.L, Zhang, K, Wang, C.C, Fan, Q, Tang, X.J, Zhang, X, Wang, K, Fu, Y, Liang, H.H. | | Deposit date: | 2023-09-13 | | Release date: | 2024-03-13 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | A c-di-GMP signaling module controls responses to iron in Pseudomonas aeruginosa.
Nat Commun, 15, 2024
|
|
4CKH
 
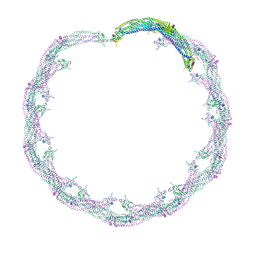 | | Helical reconstruction of ACAP1(BAR-PH domain) decorated membrane tubules by cryo-electron microscopy | | Descriptor: | ARF-GAP WITH COILED-COIL, ANK REPEAT AND PH DOMAIN-CONTAINING PROTEIN 1 | | Authors: | Pang, X.Y, Fan, J, Zhang, Y, Zhang, K, Gao, B.Q, Ma, J, Li, J, Deng, Y.C, Zhou, Q.J, Hsu, V, Sun, F. | | Deposit date: | 2014-01-06 | | Release date: | 2014-10-15 | | Last modified: | 2024-05-08 | | Method: | ELECTRON MICROSCOPY (17 Å) | | Cite: | A Ph Domain in Acap1 Possesses Key Features of the Bar Domain in Promoting Membrane Curvature.
Dev.Cell, 31, 2014
|
|
4CKG
 
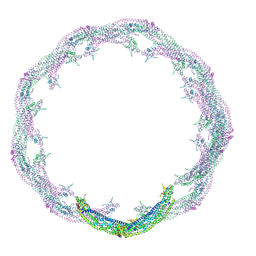 | | Helical reconstruction of ACAP1(BAR-PH domain) decorated membrane tubules by cryo-electron microscopy | | Descriptor: | ARF-GAP WITH COILED-COIL, ANK REPEAT AND PH DOMAIN-CONTAINING PROTEIN 1 | | Authors: | Pang, X.Y, Fan, J, Zhang, Y, Zhang, K, Gao, B.Q, Ma, J, Li, J, Deng, Y.C, Zhou, Q.J, Hsu, V, Sun, F. | | Deposit date: | 2014-01-06 | | Release date: | 2014-10-15 | | Last modified: | 2024-05-08 | | Method: | ELECTRON MICROSCOPY (15 Å) | | Cite: | A Ph Domain in Acap1 Possesses Key Features of the Bar Domain in Promoting Membrane Curvature.
Dev.Cell, 31, 2014
|
|
8D1T
 
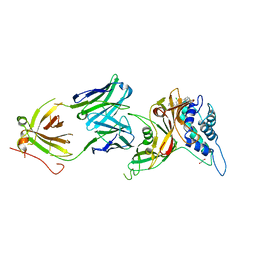 | | Crystal structure of human USP30 in complex with a covalent inhibitor 552 and a Fab | | Descriptor: | (1R,2R,4S,7E)-7-[amino(sulfanyl)methylidene]-2-{[(1P)-3-chloro-3'-(1-cyanocyclopropyl)[1,1'-biphenyl]-4-carbonyl]amino}-7-azabicyclo[2.2.1]heptan-7-ium, 1,2-ETHANEDIOL, Ubiquitin carboxyl-terminal hydrolase 30, ... | | Authors: | Song, X, Butler, J, Li, C, Zhang, K, Zhang, D, Hao, Y. | | Deposit date: | 2022-05-27 | | Release date: | 2023-02-01 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.94 Å) | | Cite: | TBD
To Be Published
|
|
6WLL
 
 | | Apo F. nucleatum glycine riboswitch models, 10.0 Angstrom resolution | | Descriptor: | RNA (171-MER) | | Authors: | Kappel, K, Zhang, K, Su, Z, Watkins, A.M, Kladwang, W, Li, S, Pintilie, G, Topkar, V.V, Rangan, R, Zheludev, I.N, Yesselman, J.D, Chiu, W, Das, R. | | Deposit date: | 2020-04-20 | | Release date: | 2020-07-08 | | Last modified: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (10 Å) | | Cite: | Accelerated cryo-EM-guided determination of three-dimensional RNA-only structures.
Nat.Methods, 17, 2020
|
|
6WLJ
 
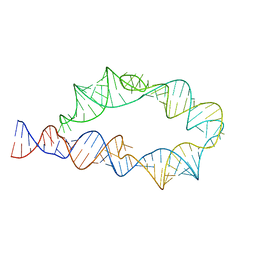 | | ATP-TTR-3 with AMP models, 9.6 Angstrom resolution | | Descriptor: | RNA (130-MER) | | Authors: | Kappel, K, Zhang, K, Su, Z, Watkins, A.M, Kladwang, W, Li, S, Pintilie, G, Topkar, V.V, Rangan, R, Zheludev, I.N, Yesselman, J.D, Chiu, W, Das, R. | | Deposit date: | 2020-04-20 | | Release date: | 2020-07-08 | | Last modified: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (9.6 Å) | | Cite: | Accelerated cryo-EM-guided determination of three-dimensional RNA-only structures.
Nat.Methods, 17, 2020
|
|
6WLT
 
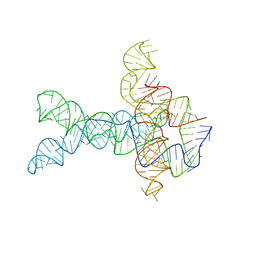 | | Apo V. cholerae glycine riboswitch models, 4.8 Angstrom resolution | | Descriptor: | RNA (231-MER) | | Authors: | Kappel, K, Zhang, K, Su, Z, Watkins, A.M, Kladwang, W, Li, S, Pintilie, G, Topkar, V.V, Rangan, R, Zheludev, I.N, Yesselman, J.D, Chiu, W, Das, R. | | Deposit date: | 2020-04-20 | | Release date: | 2020-07-08 | | Last modified: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (4.8 Å) | | Cite: | Accelerated cryo-EM-guided determination of three-dimensional RNA-only structures.
Nat.Methods, 17, 2020
|
|
6WLK
 
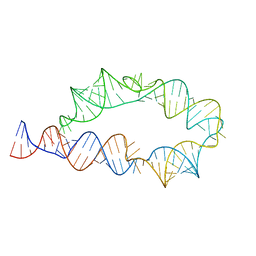 | | Apo ATP-TTR-3 models, 10.0 Angstrom resolution | | Descriptor: | RNA (130-MER) | | Authors: | Kappel, K, Zhang, K, Su, Z, Watkins, A.M, Kladwang, W, Li, S, Pintilie, G, Topkar, V.V, Rangan, R, Zheludev, I.N, Yesselman, J.D, Chiu, W, Das, R. | | Deposit date: | 2020-04-20 | | Release date: | 2020-07-08 | | Last modified: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (10 Å) | | Cite: | Accelerated cryo-EM-guided determination of three-dimensional RNA-only structures.
Nat.Methods, 17, 2020
|
|
6WLQ
 
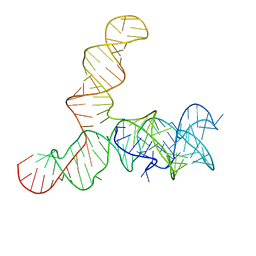 | | Apo SAM-IV riboswitch models, 4.7 Angstrom resolution | | Descriptor: | RNA (119-MER) | | Authors: | Kappel, K, Zhang, K, Su, Z, Watkins, A.M, Kladwang, W, Li, S, Pintilie, G, Topkar, V.V, Rangan, R, Zheludev, I.N, Yesselman, J.D, Chiu, W, Das, R. | | Deposit date: | 2020-04-20 | | Release date: | 2020-07-08 | | Last modified: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (4.7 Å) | | Cite: | Accelerated cryo-EM-guided determination of three-dimensional RNA-only structures.
Nat.Methods, 17, 2020
|
|
