6O52
 
 | |
4YF6
 
 | | Crystal structure of oxidised Rv1284 | | Descriptor: | Beta-carbonic anhydrase 1, CHLORIDE ION, MAGNESIUM ION, ... | | Authors: | Hofmann, A. | | Deposit date: | 2015-02-25 | | Release date: | 2015-05-06 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (3.002 Å) | | Cite: | Chemical probing suggests redox-regulation of the carbonic anhydrase activity of mycobacterial Rv1284.
Febs J., 282, 2015
|
|
4YF4
 
 | |
6O4X
 
 | | Binary complex of native hAChE with 9-aminoacridine | | Descriptor: | 9-AMINOACRIDINE, Acetylcholinesterase, GLYCEROL, ... | | Authors: | Gerlits, O, Kovalevsky, A, Radic, Z. | | Deposit date: | 2019-03-01 | | Release date: | 2019-06-19 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | A new crystal form of human acetylcholinesterase for exploratory room-temperature crystallography studies.
Chem.Biol.Interact., 309, 2019
|
|
6O5S
 
 | |
6O5R
 
 | |
6O50
 
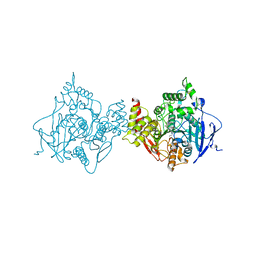 | | Binary complex of native hAChE with BW284c51 | | Descriptor: | 4-(5-{4-[DIMETHYL(PROP-2-ENYL)AMMONIO]PHENYL}-3-OXOPENTYL)-N,N-DIMETHYL-N-PROP-2-ENYLBENZENAMINIUM, Acetylcholinesterase, GLYCEROL, ... | | Authors: | Gerlits, O, Kovalevsky, A, Radic, Z. | | Deposit date: | 2019-03-01 | | Release date: | 2019-06-19 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.352 Å) | | Cite: | A new crystal form of human acetylcholinesterase for exploratory room-temperature crystallography studies.
Chem.Biol.Interact., 309, 2019
|
|
4YF5
 
 | |
3C84
 
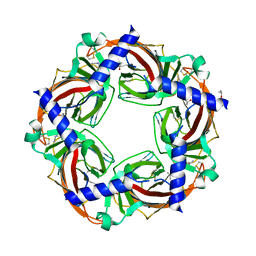 | | Crystal structure of a complex of AChBP from aplysia californica and the neonicotinoid thiacloprid | | Descriptor: | ISOPROPYL ALCOHOL, MAGNESIUM ION, Soluble acetylcholine receptor, ... | | Authors: | Talley, T.T, Harel, M, Hibbs, R.E, Tomizawa, M, Casida, J.E, Taylor, P.W. | | Deposit date: | 2008-02-08 | | Release date: | 2008-05-20 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.94 Å) | | Cite: | Atomic interactions of neonicotinoid agonists with AChBP: molecular recognition of the distinctive electronegative pharmacophore.
Proc.Natl.Acad.Sci.Usa, 105, 2008
|
|
3SH1
 
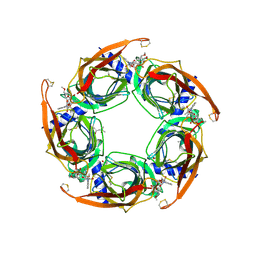 | | Ac-AChBP ligand binding domain mutated to human alpha-7 nAChR | | Descriptor: | (4R)-2-METHYLPENTANE-2,4-DIOL, (4S)-2-METHYL-2,4-PENTANEDIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Nemecz, A, Taylor, P.W. | | Deposit date: | 2011-06-15 | | Release date: | 2011-10-26 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Creating an alpha-7 nicotinic acetylcholine recognition domain from the
acetylcholine binding protein: crystallographic and ligand selectivity analyses
J.Biol.Chem., 286, 2011
|
|
3SIO
 
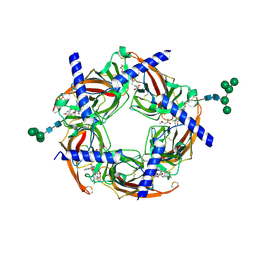 | | Ac-AChBP ligand binding domain (not including beta 9-10 linker) mutated to human alpha-7 nAChR | | Descriptor: | (4R)-2-METHYLPENTANE-2,4-DIOL, (4S)-2-METHYL-2,4-PENTANEDIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Nemecz, A, Taylor, P.W. | | Deposit date: | 2011-06-19 | | Release date: | 2011-10-26 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.32 Å) | | Cite: | Creating an alpha-7 nicotinic acetylcholine recognition domain from the
acetylcholine binding protein: crystallographic and ligand selectivity analyses
J.Biol.Chem., 286, 2011
|
|
3T4M
 
 | | Ac-AChBP ligand binding domain mutated to human alpha-7 nAChR (intermediate) | | Descriptor: | (4R)-2-METHYLPENTANE-2,4-DIOL, (4S)-2-METHYL-2,4-PENTANEDIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Nemecz, A, Taylor, P.W. | | Deposit date: | 2011-07-26 | | Release date: | 2011-10-26 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Creating an alpha-7 nicotinic acetylcholine recognition domain from the
acetylcholine binding protein: crystallographic and ligand selectivity analyses
J.Biol.Chem., 286, 2011
|
|
4NUK
 
 | | Crystal structure of nickel-bound Na-ASP-2 | | Descriptor: | Ancylostoma secreted protein 2, NICKEL (II) ION | | Authors: | Hofmann, A. | | Deposit date: | 2013-12-03 | | Release date: | 2014-05-14 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Probing the equatorial groove of the hookworm protein and vaccine candidate antigen, Na-ASP-2.
Int.J.Biochem.Cell Biol., 50, 2014
|
|
4NUI
 
 | | Crystal structure of cobalt-bound Na-ASP-2 | | Descriptor: | Ancylostoma secreted protein 2, COBALT (II) ION | | Authors: | Hofmann, A. | | Deposit date: | 2013-12-03 | | Release date: | 2014-05-14 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Probing the equatorial groove of the hookworm protein and vaccine candidate antigen, Na-ASP-2.
Int.J.Biochem.Cell Biol., 50, 2014
|
|
4NUN
 
 | | Crystal structure of copper-bound Na-ASP-2 | | Descriptor: | Ancylostoma secreted protein 2, COPPER (II) ION | | Authors: | Hofmann, A. | | Deposit date: | 2013-12-03 | | Release date: | 2014-05-14 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Probing the equatorial groove of the hookworm protein and vaccine candidate antigen, Na-ASP-2.
Int.J.Biochem.Cell Biol., 50, 2014
|
|
4NUO
 
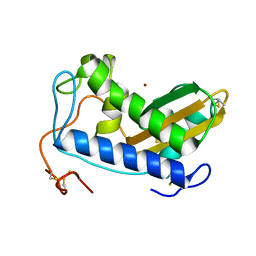 | | Crystal structure of zinc-bound Na-ASP-2 | | Descriptor: | Ancylostoma secreted protein 2, ZINC ION | | Authors: | Hofmann, A. | | Deposit date: | 2013-12-03 | | Release date: | 2014-05-14 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Probing the equatorial groove of the hookworm protein and vaccine candidate antigen, Na-ASP-2.
Int.J.Biochem.Cell Biol., 50, 2014
|
|
6U34
 
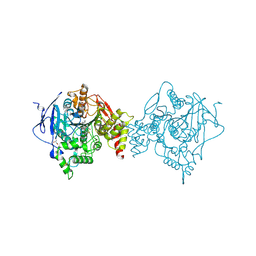 | | Binary complex of native hAChE with oxime reactivator RS194B | | Descriptor: | (2E)-N-[2-(azepan-1-yl)ethyl]-2-(hydroxyimino)acetamide, Acetylcholinesterase, GLYCEROL, ... | | Authors: | Kovalevsky, A, Gerlits, O, Radic, Z. | | Deposit date: | 2019-08-21 | | Release date: | 2020-02-12 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Rational design, synthesis, and evaluation of uncharged, "smart" bis-oxime antidotes of organophosphate-inhibited human acetylcholinesterase.
J.Biol.Chem., 295, 2020
|
|
6U37
 
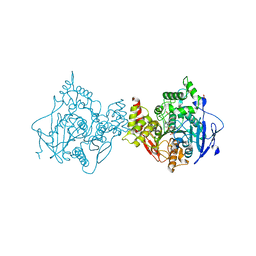 | | Structure of VX-phosphonylated hAChE in complex with oxime reactivator RS194B | | Descriptor: | (2E)-N-[2-(azepan-1-yl)ethyl]-2-(hydroxyimino)acetamide, Acetylcholinesterase, GLYCEROL, ... | | Authors: | Kovalevsky, A, Gerlits, O, Radic, Z. | | Deposit date: | 2019-08-21 | | Release date: | 2020-02-12 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Rational design, synthesis, and evaluation of uncharged, "smart" bis-oxime antidotes of organophosphate-inhibited human acetylcholinesterase.
J.Biol.Chem., 295, 2020
|
|
6U3P
 
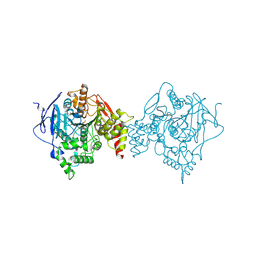 | | Binary complex of native hAChE with oxime reactivator LG-703 | | Descriptor: | (2E,2'E)-N,N'-[1,4-diazepane-1,4-diyldi(ethane-2,1-diyl)]bis[2-(hydroxyimino)acetamide], Acetylcholinesterase, DIMETHYL SULFOXIDE | | Authors: | Kovalevsky, A, Gerlits, O, Radic, Z. | | Deposit date: | 2019-08-22 | | Release date: | 2020-02-12 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Rational design, synthesis, and evaluation of uncharged, "smart" bis-oxime antidotes of organophosphate-inhibited human acetylcholinesterase.
J.Biol.Chem., 295, 2020
|
|
2IGU
 
 | | Deamidated analogue of ImI Conotoxin | | Descriptor: | Alpha-conotoxin ImI | | Authors: | Kini, R.M, Kang, T.S. | | Deposit date: | 2006-09-25 | | Release date: | 2007-08-14 | | Last modified: | 2022-03-09 | | Method: | SOLUTION NMR | | Cite: | Protein folding determinants: structural features determining alternative disulfide pairing in alpha- and chi/lambda-conotoxins
Biochemistry, 46, 2007
|
|
2IH6
 
 | | Pro6 variant of CMrVIA conotoxin | | Descriptor: | Lambda-conotoxin CMrVIA | | Authors: | Kini, R.M, Kang, T.S. | | Deposit date: | 2006-09-26 | | Release date: | 2007-08-14 | | Last modified: | 2020-06-24 | | Method: | SOLUTION NMR | | Cite: | Protein folding determinants: structural features determining alternative disulfide pairing in alpha- and chi/lambda-conotoxins
Biochemistry, 46, 2007
|
|
2IHA
 
 | | Amidated variant of CMrVIA conotoxin | | Descriptor: | Lambda-conotoxin CMrVIA | | Authors: | Kini, R.M, Kang, T.S. | | Deposit date: | 2006-09-26 | | Release date: | 2007-08-14 | | Last modified: | 2020-06-24 | | Method: | SOLUTION NMR | | Cite: | Protein folding determinants: structural features determining alternative disulfide pairing in alpha- and chi/lambda-conotoxins
Biochemistry, 46, 2007
|
|
2IFI
 
 | | Ala6 Variant of ImI Conotoxin | | Descriptor: | Alpha-conotoxin ImI | | Authors: | Kini, R.M, Kang, T.S. | | Deposit date: | 2006-09-21 | | Release date: | 2007-08-14 | | Last modified: | 2020-06-24 | | Method: | SOLUTION NMR | | Cite: | Protein folding determinants: structural features determining alternative disulfide pairing in alpha- and chi/lambda-conotoxins
Biochemistry, 46, 2007
|
|
2IFZ
 
 | | Lys6 Variant of ImI Conotoxin | | Descriptor: | Alpha-conotoxin ImI | | Authors: | Kini, R.M, Kang, T.S. | | Deposit date: | 2006-09-22 | | Release date: | 2007-08-14 | | Last modified: | 2020-06-24 | | Method: | SOLUTION NMR | | Cite: | Protein folding determinants: structural features determining alternative disulfide pairing in alpha- and chi/lambda-conotoxins
Biochemistry, 46, 2007
|
|
2IH7
 
 | | Amidated Pro6 Analogue of CMrVIA conotoxin | | Descriptor: | Lambda-conotoxin CMrVIA | | Authors: | Kini, R.M, Kang, T.S. | | Deposit date: | 2006-09-26 | | Release date: | 2007-08-14 | | Last modified: | 2020-06-24 | | Method: | SOLUTION NMR | | Cite: | Protein folding determinants: structural features determining alternative disulfide pairing in alpha- and chi/lambda-conotoxins
Biochemistry, 46, 2007
|
|
