7D5N
 
 | | Crystal structure of inositol dehydrogenase homolog complexed with NADH and myo-inositol from Azotobacter vinelandii | | Descriptor: | 1,2,3,4,5,6-HEXAHYDROXY-CYCLOHEXANE, 1,4-DIHYDRONICOTINAMIDE ADENINE DINUCLEOTIDE, Oxidoreductase | | Authors: | Fukano, K, Ono, T, Suzuki, M, Takenoya, M, Ito, S, Sasaki, Y, Yajima, S. | | Deposit date: | 2020-09-27 | | Release date: | 2021-09-29 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal structure of inositol dehydrogenase complexed with NADH and myo-inositol from Azotobacter vinelandii
To Be Published
|
|
5B1D
 
 | |
5B1F
 
 | |
5Y5N
 
 | | Crystal structure of human Sirtuin 2 in complex with a selective inhibitor | | Descriptor: | 2-[[3-(2-phenylethoxy)phenyl]amino]benzamide, NAD-dependent protein deacetylase sirtuin-2, ZINC ION | | Authors: | Mellini, P, Itoh, Y, Tsumoto, H, Li, Y, Suzuki, M, Tokuda, N, Kakizawa, T, Miura, Y, Takeuchi, J, Lahtela-Kakkonen, M, Suzuki, T. | | Deposit date: | 2017-08-09 | | Release date: | 2017-09-06 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Potent mechanism-based sirtuin-2-selective inhibition by anin situ-generated occupant of the substrate-binding site, "selectivity pocket" and NAD+-binding site.
Chem Sci, 8, 2017
|
|
1WZV
 
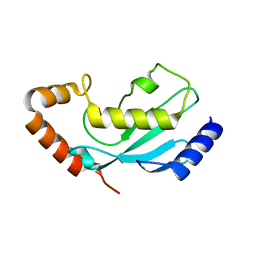 | | Crystal Structure of UbcH8 | | Descriptor: | Ubiquitin-conjugating enzyme E2 L6 | | Authors: | Mizushima, T, Suzuki, M, Teshima, N, Yamane, T, Murata, S, Tanaka, K. | | Deposit date: | 2005-03-10 | | Release date: | 2005-03-22 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystal Structure of UbcH8
To be Published
|
|
1WZW
 
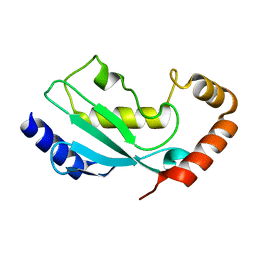 | | Crystal Structure of UbcH8 | | Descriptor: | Ubiquitin-conjugating enzyme E2 L6 | | Authors: | Mizushima, T, Suzuki, M, Teshima, N, Yamane, T, Murata, S, Tanaka, K. | | Deposit date: | 2005-03-10 | | Release date: | 2005-03-22 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Crystal Structure of UbcH8
To be Published
|
|
5B21
 
 | |
5AZ0
 
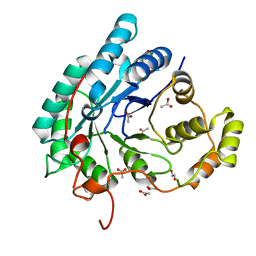 | | Crystal structure of aldo-keto reductase (AKR2E5) of the silkworm, Bombyx mori | | Descriptor: | 1,2-ETHANEDIOL, ACETATE ION, CALCIUM ION, ... | | Authors: | Yamamoto, K, Higashiura, A, Suzuki, M, Nakagawa, A. | | Deposit date: | 2015-09-15 | | Release date: | 2016-02-10 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structural characterization of an aldo-keto reductase (AKR2E5) from the silkworm Bombyx mori
Biochem.Biophys.Res.Commun., 474, 2016
|
|
5AZ1
 
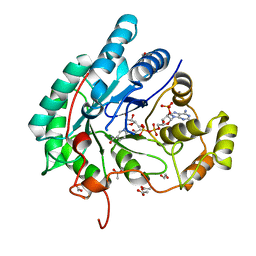 | | Crystal structure of aldo-keto reductase (AKR2E5) complexed with NADPH | | Descriptor: | 1,2-ETHANEDIOL, ACETATE ION, CALCIUM ION, ... | | Authors: | Yamamoto, K, Higashiura, A, Suzuki, M, Nakagawa, A. | | Deposit date: | 2015-09-15 | | Release date: | 2016-02-10 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structural characterization of an aldo-keto reductase (AKR2E5) from the silkworm Bombyx mori
Biochem.Biophys.Res.Commun., 474, 2016
|
|
5B22
 
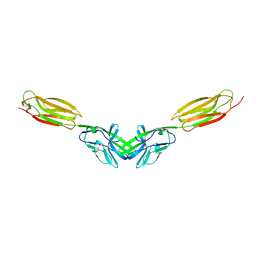 | | Dimer structure of murine Nectin-3 D1D2 | | Descriptor: | Nectin-3, beta-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose | | Authors: | Takebe, K, Sangawa, T, Katsutani, T, Narita, H, Suzuki, M. | | Deposit date: | 2015-12-28 | | Release date: | 2016-12-28 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.58 Å) | | Cite: | Dimer structure of murine Nectin-3 D1D2
To Be Published
|
|
7DDA
 
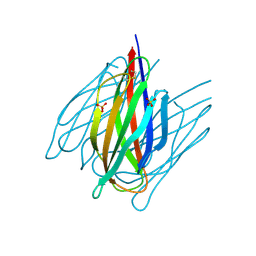 | | Envelope protein VP37 a crystal structure from White Spot Syndrome Virus | | Descriptor: | Envelope protein, SULFATE ION | | Authors: | Somsoros, W, Sangawa, T, Takebe, K, Attarataya, J, Suzuki, M, Khunrae, P. | | Deposit date: | 2020-10-28 | | Release date: | 2021-06-23 | | Method: | X-RAY DIFFRACTION (2.51 Å) | | Cite: | Crystal structure of the C-terminal domain of envelope protein VP37 from white spot syndrome virus reveals sulphate binding sites responsible for heparin binding.
J.Gen.Virol., 102, 2021
|
|
2E1A
 
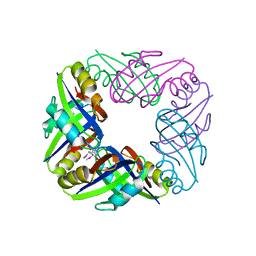 | | crystal structure of FFRP-DM1 | | Descriptor: | 75aa long hypothetical regulatory protein AsnC, SELENOMETHIONINE | | Authors: | Koike, H, Suzuki, M. | | Deposit date: | 2006-10-19 | | Release date: | 2007-09-18 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | A Structural Code for Discriminating between Transcription Signals Revealed by the Feast/Famine Regulatory Protein DM1 in Complex with Ligands
Structure, 15, 2007
|
|
2DCZ
 
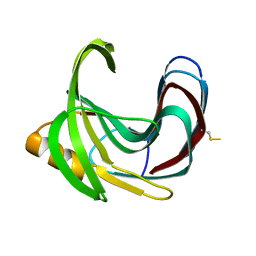 | | Thermal Stabilization of Bacillus subtilis Family-11 Xylanase By Directed Evolution | | Descriptor: | 1,4-DIETHYLENE DIOXIDE, Endo-1,4-beta-xylanase A, SULFATE ION | | Authors: | Kondo, H, Miyazaki, K, Takenouchi, M, Noro, N, Suzuki, M, Tsuda, S. | | Deposit date: | 2006-01-18 | | Release date: | 2006-02-07 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Thermal Stabilization of Bacillus subtilis Family-11 Xylanase by Directed Evolution
J.Biol.Chem., 281, 2006
|
|
2DCY
 
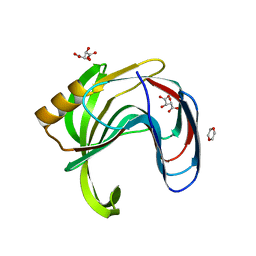 | | Crystal structure of Bacillus subtilis family-11 xylanase | | Descriptor: | 1,4-DIETHYLENE DIOXIDE, D(-)-TARTARIC ACID, Endo-1,4-beta-xylanase A, ... | | Authors: | Kondo, H, Miyazaki, K, Takenouchi, M, Noro, N, Suzuki, M, Tsuda, S. | | Deposit date: | 2006-01-18 | | Release date: | 2006-02-07 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Thermal Stabilization of Bacillus subtilis Family-11 Xylanase by Directed Evolution
J.Biol.Chem., 281, 2006
|
|
2E5A
 
 | | Crystal Structure of Bovine Lipoyltransferase in Complex with Lipoyl-AMP | | Descriptor: | 5'-O-[(R)-({5-[(3R)-1,2-DITHIOLAN-3-YL]PENTANOYL}OXY)(HYDROXY)PHOSPHORYL]ADENOSINE, ACETIC ACID, Lipoyltransferase 1, ... | | Authors: | Fujiwara, K, Hosaka, H, Matsuda, M, Suzuki, M, Nakagawa, A. | | Deposit date: | 2006-12-19 | | Release date: | 2007-09-04 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystal structure of bovine Lipoyltransferase in complex with lipoyl-AMP
J.Mol.Biol., 371, 2007
|
|
2E1C
 
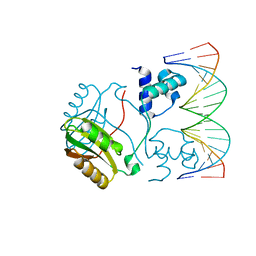 | | Structure of Putative HTH-type transcriptional regulator PH1519/DNA Complex | | Descriptor: | DNA (5'-D(*DAP*DGP*DTP*DGP*DAP*DAP*DAP*DAP*DTP*DTP*DTP*DTP*DTP*DCP*DAP*DCP*DA)-3'), DNA (5'-D(*DTP*DGP*DTP*DGP*DAP*DAP*DAP*DAP*DAP*DTP*DTP*DTP*DTP*DCP*DAP*DCP*DT)-3'), Putative HTH-type transcriptional regulator PH1519 | | Authors: | Koike, H, Suzuki, M. | | Deposit date: | 2006-10-24 | | Release date: | 2007-12-04 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Feast/Famine Regulation by Transcription Factor FL11 for the Survival of the Hyperthermophilic Archaeon Pyrococcus OT3.
Structure, 15, 2007
|
|
2D05
 
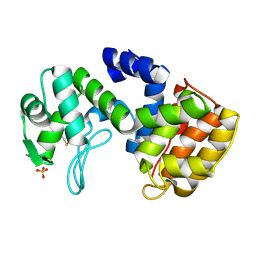 | | Chitosanase From Bacillus circulans mutant K218P | | Descriptor: | Chitosanase, SULFATE ION | | Authors: | Fukamizo, T, Amano, S, Yamaguchi, K, Yoshikawa, T, Katsumi, T, Saito, J, Suzuki, M, Miki, K, Nagata, Y, Ando, A. | | Deposit date: | 2005-07-25 | | Release date: | 2005-12-06 | | Last modified: | 2021-11-10 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Bacillus circulans MH-K1 Chitosanase: Amino Acid Residues Responsible for Substrate Binding
J.Biochem.(Tokyo), 138, 2005
|
|
2E0Z
 
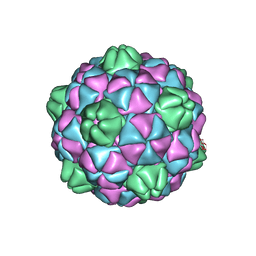 | | Crystal structure of virus-like particle from Pyrococcus furiosus | | Descriptor: | Virus-like particle | | Authors: | Akita, F, Chong, K.T, Tanaka, H, Yamashita, E, Miyazaki, N, Nakaishi, Y, Namba, K, Ono, Y, Suzuki, M, Tsukihara, T, Nakagawa, A. | | Deposit date: | 2006-10-16 | | Release date: | 2007-04-17 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (3.6 Å) | | Cite: | The Crystal Structure of a Virus-like Particle from the Hyperthermophilic Archaeon Pyrococcus furiosus Provides Insight into the Evolution of Viruses
J.Mol.Biol., 368, 2007
|
|
5WRB
 
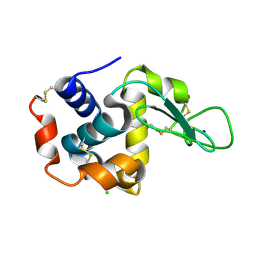 | | Crystal structure of hen egg-white lysozyme | | Descriptor: | CHLORIDE ION, Lysozyme C, SODIUM ION | | Authors: | Sugahara, M, Suzuki, M, Masuda, T, Inoue, S, Nango, E. | | Deposit date: | 2016-12-01 | | Release date: | 2017-12-20 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.013 Å) | | Cite: | Hydroxyethyl cellulose matrix applied to serial crystallography
Sci Rep, 7, 2017
|
|
5Y1A
 
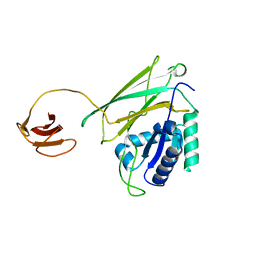 | | HBP35 of Porphyromonas gingivalis | | Descriptor: | 35 kDa hemin binding protein | | Authors: | Kakuda, S, Suzuki, M, Sato, K. | | Deposit date: | 2017-07-20 | | Release date: | 2018-07-25 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Immunoglobulin-like domains of the cargo proteins are essential for protein stability during secretion by the type IX secretion system.
Mol. Microbiol., 110, 2018
|
|
5WRC
 
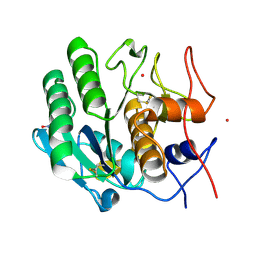 | | Crystal structure of proteinase K from Engyodontium album | | Descriptor: | NITRATE ION, PRASEODYMIUM ION, Proteinase K | | Authors: | Sugahara, M, Nakane, T, Suzuki, M, Masuda, T, Inoue, S, Numata, K. | | Deposit date: | 2016-12-01 | | Release date: | 2017-11-29 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Hydroxyethyl cellulose matrix applied to serial crystallography
Sci Rep, 7, 2017
|
|
5X9M
 
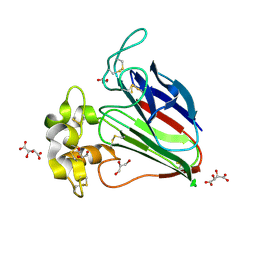 | | Structure of hyper-sweet thaumatin (D21N) | | Descriptor: | GLYCEROL, L(+)-TARTARIC ACID, Thaumatin I | | Authors: | Masuda, T, Okubo, K, Sugahara, M, Suzuki, M, Mikami, B. | | Deposit date: | 2017-03-08 | | Release date: | 2018-03-14 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (0.93 Å) | | Cite: | Subatomic structure of hyper-sweet thaumatin D21N mutant reveals the importance of flexible conformations for enhanced sweetness.
Biochimie, 157, 2019
|
|
5X9L
 
 | | Recombinant thaumatin I at 0.9 Angstrom | | Descriptor: | GLYCEROL, L(+)-TARTARIC ACID, Thaumatin I | | Authors: | Masuda, T, Okubo, K, Sugahara, M, Suzuki, M, Mikami, B. | | Deposit date: | 2017-03-08 | | Release date: | 2018-03-14 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (0.9 Å) | | Cite: | Subatomic structure of hyper-sweet thaumatin D21N mutant reveals the importance of flexible conformations for enhanced sweetness.
Biochimie, 157, 2019
|
|
8K6T
 
 | | The minor pilin structure of FctB3 in Streptococcus | | Descriptor: | FctB3, GLYCEROL | | Authors: | Takebe, K, Sangawa, T, Suzuki, M, Nakata, M. | | Deposit date: | 2023-07-25 | | Release date: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Analysis of FctB3 crystal structure and insight into its structural stabilization and pilin linkage mechanisms.
Arch.Microbiol., 206, 2023
|
|
5YYP
 
 | | Structure K137A thaumatin | | Descriptor: | GLYCEROL, L(+)-TARTARIC ACID, Preprothaumatin I | | Authors: | Masuda, T, Kigo, S, Mitsumoto, M, Ohta, K, Suzuki, M, Mikami, B, Kitabatake, N, Tani, F. | | Deposit date: | 2017-12-10 | | Release date: | 2018-03-21 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.01 Å) | | Cite: | Positive Charges on the Surface of Thaumatin Are Crucial for the Multi-Point Interaction with the Sweet Receptor.
Front Mol Biosci, 5, 2018
|
|
